
Upregulation of PAIP1 promotes the gallbladder tumorigenesis through regulating PLK1 level
Introduction
Gallbladder cancer (GBC) is a rare biliary tract malignancy and ranks fifth in tumors of the digestive tract worldwide (1). The 5-year survival rate of GBC patients is less than 5%, likely because of its early metastasis, late diagnosis, as well as poor prognosis (2,3). Currently, radical surgery is the only powerful curative treatment option for GBC (3), whereas chemotherapeutic approaches can only extend overall survival by a few months (4). Furthermore, due to its late and nonspecific symptoms, GBC is commonly diagnosed at advanced stages, which makes treatment choices are limited and prognosis is poor. Therefore, it is of paramount importance to explore the molecular mechanisms and potential therapeutic targets for GBC.
PolyA-binding protein interacting protein 1 (PAIP1) is encoded by the PAIP1 gene, which contains 2 interacting motifs (PAM1 and PAM2) for binding the PolyA-binding protein (PABP) (5,6). While PABP forms a complex with the eukaryotic initiation factor 4G (eIF4G) to regulate messenger RNA (mRNA) circularization, the association of PAIP1 and PABP further promotes PABP’s activity on translation initiation (5,7). Specifically, PAIP1 simultaneously interacts with PABP along with the translation initiation factors, eIF4 and eIF3, to facilitate 5’ cap-dependent translation (5,7). Furthermore, ribosomal protein S6 kinase 1 and 2 (S6K1/2) are capable of phosphorylating eIF3 to enhance PAIP1-eIF3 interaction and translation initiation in a nutrient-dependent manner (8). Meanwhile, addition of the inhibitors, rapamycin and PP242, which target the mechanistic target of rapamycin complex 1 (mTORC1), can disrupt their association and reduce the translation efficiency (8). In mice, PAIP1 specifically binds to YBX2, a RNA-binding protein that participates in mRNA storage, which stimulates translation of spermatogenic mRNAs during spermiogenesis (9). The results indicate a critical role of PAIP1 in male germ cell development. Recently, it has been reported that the eukaryotic polypeptide chain release factor, eRF3, competes with PAIP1 in binding PABP at the same domain, which attenuates PAIP1’s activity towards PABP and promotes translation termination via the PABP-eRF3 complex (10). In addition, PAIP1 forms a multiprotein complex to impede RNA deadenylation and protect mRNA decay (11). These findings clearly suggest PAIP1 plays important roles in mRNA translation and stability.
Intriguingly, an abnormally high expression of PAIP1 has been demonstrated to be linked with poor overall survival in various human cancers, including breast, gastric, liver, pancreatic, and lung cancers (12-16). For example, using different tissue-derived cell lines, Lin group has shown that overexpression of PAIP1 promotes cell proliferation, metastasis, and angiogenesis, whereas knockdown of PAIP1 inhibits these phenotypes, in both breast and pancreatic cancers, suggesting its pathological roles in tumorigenesis (12). Although PAIP1 may participate in lung adenocarcinoma via the AKT/GSK3β pathway (13) or in pancreatic cancer via AKT pathway (12), whether PAIP1 takes part in GBC remains unknown, and thus exploring the physiological function of PAIP1 and its potential relevance in GBC may be a fruitful avenue of research.
In this study, we discovered that upregulated PAIP1 expression was positively correlated with GBC. We further observed that knockdown of PAIP1 in two gallbladder cell lines inhibited cell proliferation, reduced colony formation, and induced apoptosis. At the animal level, we showed that knockdown of PAIP1 remarkably reduced xenograft tumor growth, reinforcing a critical role of PAIP1 in gallbladder oncogenesis. Mechanistically, using gene expression profiling microarray analysis we found that stable silencing of PAIP1 altered various gene expressions, including many genes that regulate cell cycle progression. Finally, found that PLK1 kinase, a key regulator of cell cycle progression, is regulated at the transcription and protein levels by PAIP1, and that rescue of PAIP1 level can restore the PLK1 level and activate cell growth. Therefore, we propose that PAIP1 likely contributes to GBC progression through regulating PLK1 and thus may be a clinical prognostic biomarker of GBC. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/atm-21-2417).
Methods
Cell lines and culture
The human GBC cell lines, GBC-SD and NOZ, used in this study were purchased from the Cell Bank of the Chinese Academy of Science (Shanghai, China) and the Fenghui Bio Company (Changsha, China), respectively. Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 U/mL penicillin/streptomycin in a humidified incubator with 5% CO2 and at 37 °C.
RNA interference and vectors
Small interfering RNAs (siRNAs) that specifically target human PAIP1 were purchased from GenePharma (Shanghai, China). The siRNA sequences were listed in Table S1. The vectors, pCS2-3xFlag-PAIP1 and pCS2-3xHA-PAIP1, were cloned from complement DNA (cDNA) extracted from HEK 293T cells, and the DNA sequences were validated by sequencing (TsingKe Company, China). Cells were cultured on 6-well plates to ~80% confluence and transfected with siRNAs or negative control using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions. The shPAIP1 and scrambled short hairpin control (shCtrl) lentivirus vectors were purchased from GeneChem (Shanghai, China). The shPAIP1 sequences in the vector were listed in Table S1. Stably shPAIP1-transfected cells were collected though treatment with puromycin (1 μg/mL, Solarbio, China).
Collection of GBC microarray data
Microarray data sets (GSE139682, GSE132223, and GSE139682) available online were analyzed using the Gene Expression Omnibus (GEO) of the National Center for Biotechnology Information (NCBI). The cholangiocarcinoma data set was obtained from The Cancer Genome Atlas (TCGA).
Cell proliferation and cell cycle analysis
For cell growth assays, 2×103 cells per well were seeded into 96-well plates, and 3 biological wells per group were analyzed. Cell numbers in each well were evaluated using a cell counting kit-8 (CCK-8; Engreen Biosystem, Ltd., Auckland, New Zealand) after 5 days. Briefly, 10 microliters of CCK-8 reagent was added to each well, and the plate was then incubated at 37 °C for 2 hours. Subsequently, the absorbance at 490 nm was measured in each well with a spectrophotometer. For cell cycle progression analysis, the indicated cells were fixed with precooled 70% ethanol overnight at 4 °C and incubated in staining solution (5 U/mL RNase A and 10 μg/mL propidium iodide) at 37 °C for 30 min. The cell cycle distribution was assayed by flow cytometry on a FACSCalibur (BD Biosciences, USA), and the data were analyzed with FlowJo software.
Western blotting
Western blotting was performed as described previously (17). In brief, cells were collected and lysed by RIPA buffer with a protease inhibitor cocktail (Bimaker, China). Equal amounts of protein were electrophoresed on sodium dodecyl sulfate polyacrylamide gel electrophoresis gels (SDS-PAGE) and then transferred to polyvinylidene difluoride membranes (Millipore, MA, USA). After that, the membranes were blocked in buffer [5% free-fat milk in tris-buffered saline with Tween20 (TBST)] before being probed with the primary antibodies listed in the Table S2.
Apoptotic assay
shCtrl or shPAIP1 cells were trypsinized in the logarithmic growth phase for 1 hour. Both adherent and floating cells were collected and washed 3 times with ice-cold phosphate-buffered saline. Subsequently, flow cytometry was used to detect apoptotic cells double stained by Annexin V-APC staining (88-8007; eBioscience, San Diego, CA, USA) and propidium iodide within 15 minutes according to the manufacturer’s instructions. Each experiment was performed in triplicate.
Colony formation assay
Cells were seeded at a density of 1×103 cells in six-well plates and incubated at 37 °C for 10 d. The cell colonies were rinsed with PBS, then fixed with methanol for 10 min, and stained with methylthionine chloride. The number of colonies with more than 50 cells was counted with phase contrast imaging using a microscope (XDS-100, CAIKON, China).
Caspase 3/7 assay
Caspase 3/7 assay was performed according to the manufacturer’s protocol for cells cultured in a 96-well plate (G8091; Promega, Madison, WI, USA).
Transwell assays
NOZ cells were seeded into D60 plates and then cultured to 50% confluence with a concentration of 1×106 cells. Cells were transfected with 200 pmol siRNAs targeting PAIP1 using lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions. Cells were transfected for 48 hours and seeded in 24-well plates (5,000 cells each) with 8-µm pore sizes (BD Bioscience, USA) for the migration assay. The invasion assay was analyzed using the same transwell inserts coated with Matrigel. Cells were seeded into the upper insert in serum-free media, and complete medium containing 10% FBS was added to the bottom wells incubated at 37 °C for 36 hours. Cells on the lower filter surface were fixed with 4% paraformaldehyde for 30 min, and stained with Giemsa for 15 min. The number of cells in three fields from three independent wells was counted.
Tumor xenograft experiments
Female BALB/c nude mice (4 weeks old) were obtained from LingChang Biotech (Shanghai, China) and raised following a regular procedure. NOZ cells (4×106/mL) transfected with the luciferase (Luc)-labeled shPAIP1 or shCtrl lentivirus construct were injected subcutaneously into the right flanks of BALB/C 10 nude mice. Tumor growth was measured following the indicated time points, and tumor sizes were calculated by the following formula: volume (cm3) = 0.5 × length × width2. After 4 weeks, 10 µL/g of D-Luciferin (15 mg/mL) was injected intraperitoneally into these mice before they were sacrificed, after which the tumors were collected and weighed. The fluorescence in each mouse was detected using the Lumina LT imaging system (PerkinElmer, Waltham, MA, USA). All procedures were conducted in accordance with the Guidelines for the Care and Use of Laboratory. Animal experiments were approved by the Ethics Committee of the Tongji Medical College, Huazhong University of Science and Technology (No. GSGC0162883).
IHC staining
A total of 22 GBC tissues were collected from patients in Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China. Tissue histosection slides were de-paraffinized and rehydrated following the protocols described previously (18). The slides were incubated with rabbit anti-human polyclonal antibodies against PAIP1 (1:200 dilution) and Ki-67 (1:100 dilution) at 4 °C overnight and then probed with biotin-conjugated goat anti–rabbit secondary antibody at room temperature for 30 min. Slides were incubated with streptavidin-peroxidase complex. The peroxidase reaction was developed with 3, 3’-diaminobenzidine and counterstained with Mayer’s hematoxylin. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Tongji Medical College, Huazhong University of Science and Technology (LLHBCH2019LW-003), and informed consent was provided by all participants.
The intensity score was determined by evaluating staining intensity of positive staining as described previously (19). Samples with histoscore of more than 4 were considered to be high, while those with a score less than 4 were considered to be low.
Coimmunoprecipitation (co-IP) experiment
Co-IP experiments were performed following the procedures as described before (20). The resulting immune-precipitates were subjected to immunoblotting with the indicated antibodies. To construct the pCS2-3xFlag-PLK1-TurboID plasmid, the pCS2-3xFlag-PLK1 reported previously (21) was digested with XbaI and XhoI restriction enzymes. The DNA fragment of TurboID was cloned into this digested vector by a HieffClone Plus multi One-Step Cloning Kit (10912ES10, Yesen, China). The TurboID fragment was amplified by PCR from a commercial gene (USA126049; Addgene, Watertown, MA, USA). The sequences of the primers were listed in Table S1. For biotin labeling of transiently transfected cells, the pCS2-3xFlag-PLK1-TurboID construct was transfected into HEK 293T cells, and biotin at a final concentration of 50 µM was added just 6 hours before cells were harvested (22). To isolate the biotinylated proteins that bind with PLK1, cells lysates were denatured and incubated with NeutrAvidin agarose (29204; Thermo Fisher Scientific, USA) at 4 °C for 4 hours with rotation. After being washed with IP buffer 3 times, the resins were subjected to run gel and immunoblot with the indicated antibodies.
Human gene expression array
The total RNA sample was analyzed by Agilent 2100 using GeneChip 3’ in vitro translation Express Kit (Affymetrix, USA). The Affymetrix gene expression profiling microarray chip was treated as described before (20), and scanned to obtain images and raw data. A differentially expressed gene list is available in Table S1.
Statistical analysis
Data are presented as mean ± standard deviation (SD) from three experimental replicates. The Student’s t-test was used to analyze the differences between groups using GraphPad Prism 5. Statistical differences between two groups were analyzed using an independent Student’s t-test (2-tailed). A P value less than 0.05 was considered to be statistically significant. For quantification of the western blot signals, ImageJ software was used to measure the relative intensity of each band.
Results
PAIP1 expression was upregulated in GBC tissues
To investigate whether PAIP1 plays a pathologic role in GBC, relative mRNA expression levels of PAIP1 in normal tissue vs. tumor tissues in two different data sets from the GEO were analyzed. We found that PAIP1 expression levels were significantly upregulated in GBC tissues compared to normal tissues in both data sets (Figure 1A,B). It has been illustrated that GBC together with cholangiocarcinoma (CCA) constitutes a group of biliary tract cancers (19,23). To clarify whether PAIP1 is only upregulated in GBC tissues, we further analyzed expression levels of PAIP1 in CCA tissues obtained from the TCGA database. We found that PAIP1 was markedly upregulated in CCA, suggesting PAIP1’s broader effect in biliary tract cancers (Figure 1C). To confirm whether higher mRNA levels of PAIP1 are correlated with higher protein levels of PAIP1 in GBC tissues, we assessed PAIP1 protein levels in 22 GBC tissues by IHC staining (Figure 1D). Based on the histoscores, PAIP1 protein was assessed as having a higher expression in most GBC tissues (15 out of 22 gallbladder tumor tissue samples), which confirmed that PAIP1 expression is upregulated in GBC tissues (Figure 1E).
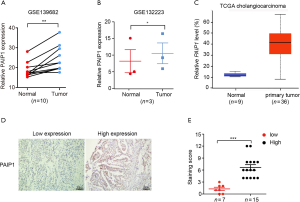
Stable silencing of PAIP1 suppressed cell proliferation, migration, and invasion
Next, two different gall bladder-derived cancer cell lines, NOZ and GBC-SD cells, were used to investigate the causal role of PAIP1 in GBC progression. Lentivirus vector-based shRNA construct bearing a specific sequence targeting PAIP1 gene (shPAIP1) was generated and transfected into NOZ and GBC-SD cells with shCtrl serving as a control. After virus-infected cells treated with puromycin were harvested, the PAIP1 knockdown efficiency was validated by qPCR analysis and western blotting. We observed that mRNA levels of PAIP1 were significantly decreased in both cell lines upon shRNA treatment (Figure 2A, Figure S1A), and PAIP1 protein levels were also decreased accordingly (Figure 2B, supplemental Figure S1B). As expected, knockdown of PAIP1 significantly retarded cell growth, as assessed by the counting of cell numbers over time (Figure 2C, Figure S1C). Cell proliferation assays revealed a suppressed capacity of cell proliferation in PAIP1-silenced cells as compared with controls (Figure 2D, Figure S1D). Furthermore, colony formation assays also showed decreased cell colony numbers in cells expressing shPAIP1 than in cells expressing shCtrl, regardless of which gallbladder cell lines were used (Figure 2E,F, Figure S1E,F). Meanwhile, transwell assays were performed to examine the effect of PAIP1 in GBC cell migration and invasion. We found that knockdown of PAIP1 markedly inhibited cell migration and invasion abilities (Figure 2G,H). Altogether, these data support that PAIP1 plays a role in GBC progression, migration and invasion.
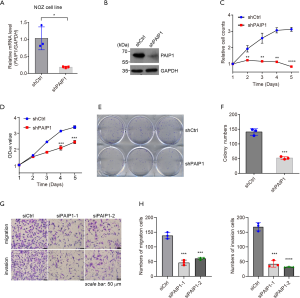
Reduction of PAIP1 level promoted gallbladder cell apoptosis and caused cell cycle arrest
Since knockdown of PAIP1 suppresses cell proliferation, we wanted to know whether this results from induced apoptosis of living cells or the arrest of cell cycle progression. Toward this end, annexin-V marked apoptotic cells were counted by flow cytometry, in which annexin-V specifically bound to phosphatidylserine exposed on the outer cellular membrane in dead cells (24). From experiments performed in triplicate, we observed that knockdown of PAIP1 increased the cell population staining with annexin-V (Figure 3A, Figure S2A) and facilitated cell apoptosis (Figure 3B, Figure S2B) in NOZ and GBC-SD cells, respectively. Caspases, including caspase-3 and caspase-7, are crucial for final apoptotic execution, and caspases become cleaved upon activation (25). Thus, a sensitive, single-step method based on bioluminescence protease reaction was carried out using peptide-conjugated aminoluciferin as the protease substrate and a firefly luciferase that has been molecularly evolved for increased stability (26). Consistently, an increased portion of apoptotic cells were observed upon knockdown of PAIP1 (Figure 3C, Figure S2C). To rule out the possibility that induced apoptosis in cells with stable silencing of PAIP1 might have been an off-target effect, 2 siRNA oligos were transiently transfected into NOZ cells, and the protein levels of caspases were examined. We found that both siRNAs targeting PAIP1 genes induced caspase-3 and caspase-7 activation as confirmed by the increase in protein levels and accumulation of cleaved caspase-3 forms, which indicated apoptotic cell progress had occurred (Figure 3D,E). Additionally, flow cytometry analysis demonstrated that knockdown of PAIP1 caused cell cycle arrest, but somehow, with an increased G1 cell population in NOZ cells (Figure 3F) but an increased G2/M population in GBC-SD cells (Figure S2D). Thus, our data suggest that the suppression of cell proliferation upon knockdown of PAIP1 in GBC cells is likely due to induced apoptosis and cell cycle arrest.
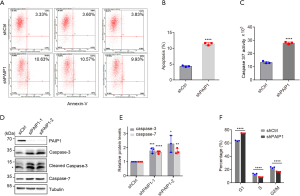
Knockdown of PAIP1 attenuated tumorigenesis in mice
To evaluate the pathological role of PAIP1 in vivo, we developed a xenograft tumor model by inoculating equal amounts of Luc-labelled shCtrl or shPAIP1 cells into the right flanks of nude mice (Figure 4A, left panel). We followed the growth of tumors over time by measuring the tumor size and weight. Compared with the control mice that exhibited a gradual increase of tumor size, the shPAIP1 mice exhibited much slower growth of tumors (Figure 4B). At 22 days post injection, the mice were sacrificed and the tumors were weighed. The tumors were consistently found to be much smaller upon knockdown of PAIP1 as compared to those of control mice (Figure 4C,D). In line with our results, upon addition of D-Luciferin into mice before they were killed, we observed stronger luminescent signals in the control mice than in the shPAIP1 mice (Figure 4A, right panel). IHC staining of mice tumor tissues revealed that knockdown of PAIP1 led to reduced expression of Ki-67, a cell proliferation marker, confirming PAIP1 level to be directly correlated with cell proliferation and tumorigenesis (Figure 4E).

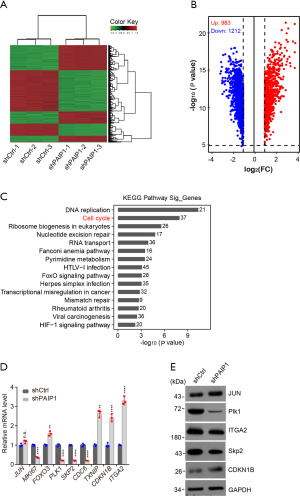
PAIP1 regulated expression of multiple genes involved in cell cycle progression
To explore the molecular mechanism of PAIP1 in gallbladder cell proliferation and tumorigenesis, an Affymetrix microarray chip was used to identify potential downstream targets of PAIP1. Subsequently, NOZ cells with shCtrl or shPAIP1 construct were harvested, and total mRNAs were purified and subjected to hybridization of the microarray chip, with the expression profiles being analyzed. We repeatedly observed that knockdown of PAIP1 markedly altered multiple gene expression levels as compared to the shCtrl cells in 3 independent samples (Figure 5A and in total online: https://cdn.amegroups.cn/static/public/atm-21-2417-1.xlsx). When we set a selected threshold with 2-fold changes and a P value of less than 0.05, 983 upregulated genes and 1212 downregulated genes were identified upon knockdown of PAIP1 relative to the shCtrl group (Figure 5B). The hierarchical clustering analysis revealed that the differentially expressed genes were mainly enriched in DNA replication and cell cycle (Figure 5C). The expression changes of several selected genes involved in these 2 pathways were validated by qPCR analysis. We noticed that knockdown of PAIP1 significantly inhibited expression of MKI67 (marker of proliferation Ki-67), PLK1 (Polo-like kinase 1), and SKP2 (S-phase kinase-associated protein 2), and promoted expression of CDKN1B (cyclin-dependent kinase inhibitor 1B), ITGA2 (alpha 2 subunit of VLA-2 receptor), and TXNIP (thioredoxin interacting protein), which is consistent with the microarray data; meanwhile, there were no changes of expression of JUN (jun proto-oncogene), which is inconsistent with the microarray data (Figure 5D). To identify which genes were indeed affected by PAIP1, the protein levels encoded by some of these genes were examined by immunoblotting. Surprisingly, we found that PLK1 level, but not other protein levels, was dramatically decreased upon knockdown of PAIP1, which might indicate the relevance of PLK1 and PAIP1 (Figure 5E).
PAIP1 promoted gallbladder tumorigenesis possibly through regulation of PLK1 level
It has been well demonstrated that PLK1 is a crucial regulator of cell cycle progression (27,28). Elevated PLK1 levels are present in many type of cancers, including GBC, and are closely linked with poor prognosis (29,30). Consistently, we also found that PLK1 levels were higher in GBC tissues obtained from the aforementioned datasets (Figure 6A). Interestingly, PLK1 expression levels were positively correlated with PAIP1 expression levels in these tissue samples (R=0.37, P=2.9e-05; Figure 6B). We conducted immunostaining of the 22 collected human GBC tissues with the PLK1 antibody, and we found that 9 samples exhibited low expression levels, while 13 samples exhibited high expression levels of PLK1 (Figure S3A,B). Consistently, PLK1 protein levels were positively correlated with PAIP1 protein levels in the 22 GBC tissue samples (R=0.7656, P<0.001; Figure S3C). To further confirm the relationship of PLK1 and PAIP1, we examined protein levels of PLK1 in xenograft tumor tissues bearing either the shCtrl or shPAIP1 lentivirus construct. The PLK1 levels significantly were reduced in shPAIP1 samples compared with shCtrl samples (Figure 6C,D), which underscores our point that PLK1 levels are positively correlated with PAIP1 levels in both the mouse model and patient GBC tissues.

It has been reported that PAIP1 regulates mRNA translation efficiency and development through interaction with several proteins (5,9,10). To test whether PAIP1 regulates GBC cell proliferation by binding PLK1, co-IP experiments were performed. Using a TurboID technique that could efficiently identify proximal and interacting proteins in their natural cellular environment (22), we found that PLK1 was able to bind with PAIP1 in HEK 293T cells (Figure 6E). Reciprocal co-IP results further confirmed that PLK1 interacts with PAIP1 (Figure 6F,G). Finally, we wondered if reintroduction of PAIP1 in PAIP1-silenced cells could recover the reduced PLK1 level. Thus, a Flag-tagged PAIP1 construct was transfected into NOZ cells with two siPAIP1 oligos, and we found that PLK1 levels were increased upon PAIP1 level recovery (Figure 6H). Moreover, caspases activities were also rescued, indicating the importance of PAIP1 in apoptosis (Figure 6H). Collectively, the results support the supposition that PAIP1 regulates gallbladder tumors through the regulation of PLK 1.
Discussion
GBC remains an understudied disease. Although its incidence is less than 2 per 100,000 individuals worldwide, patients with GBC are predominantly diagnosed at the advanced stages with 5-year survival rates being <5% (3). In contrast to more common human tumors, evidence about the molecular changes involved in the development of GBC are less known thus far. Therefore, it is crucial to identify specific regulators that are required for gallbladder carcinogenesis. The tumor suppressor genes, including TP53, ARID1A, and SMAD4, and the oncogenes, including CDKN2A/B, KRAS, and EGFR, have been characterized with mutations or genomic alterations in GBC, and this may provide direction for clinical treatment (31,32). However, only a subset of patients is directly linked with these mutations. It is thus critical to discover other key factors functioning in GBC development and progression.
In the present study, we identified poly A-binding protein-interacting protein PAIP1 as a potential regulators involved in GBC development and progression. Elevated PAIP1 level is associated with gallbladder carcinogenesis, including GBC and CCA. Using two different GBC cell lines, we further demonstrated that reduction of PAIP1 levels in tumor cells retarded cell proliferation, colony formation, promoted apoptosis, and caused cell cycle arrest. Given that we failed to obtain cholangiocarcinoma-derived cell lines, we did not examine whether the similar phenotypes would occur upon silencing of PAIP1. However, the fact that upregulated PAIP1 levels were correlated with cholangiocarcinoma tissues may lead us to investigate whether PAIP1 is also an important regulator in CCA.
To explore the possible molecular mechanism underlying how PAIP1 affects GBC, we performed high-throughput microarray chip analysis. This revealed that PAIP1 may participate in regulating cell cycle progression. After a comprehensive survey, we found that PAIP1 significantly affect a key cell cycle regulator, PLK1, thereby activating functions in GBC development and progression. Overexpression of PLK1 has been shown to occur in a wide range of tumors, and this phenomenon has attracted considerable attention from researchers in academic and commercial pharmaceutical settings seeking to develop PLK1 inhibitors as a means of cancer treatment (29). The present study is the first of its kind to link PAIP1 with PLK1 genetically and physically, which might open a window for clinical treatment of GBC.
There are a few reasons to assume PAIP1 is a regulator in gallbladder tumors acting via PLK1. First, it has been reported that PAIP1 exerts its tumor-promoting role by facilitating cell cycle progression through regulating the expression of cyclin D1 in gastric cancer (14). Researchers have shown that PAIP1 knockdown results in decreased expression of cyclin D1, whereas overexpression of PAIP1 enhances its expression. Unfortunately, we did not find that decreased PAIP1 altered cyclin D1 expression in our microarray dataset in total online: https://cdn.amegroups.cn/static/public/atm-21-2417-1.xlsx. As PLK1 is a critical cell cycle regulator, and knockdown of PAIP1 causes cell cycle arrest, it is reasonable to assume that they are linked. Second, we found that PAIP1 significantly affected he PLK1 protein level (Figure 5E), and an elevated PAIP1 level is positively correlated with higher PLK1 level in gallbladder tumors. Since PLK1 inhibition has been shown to cause cell cycle block and apoptosis, PLK1 inhibitors may be a potential cancer therapy for treatment of GBC.
However, how PAIP1 regulates PLK1 is still unclear, and several possible interactions underlying this relationship may be explored in future research. Given that PAIP1 interacts with the polyA-interacting protein PABP to promote translational efficiency, it is likely that PAIP1 affects the mRNA translation or mRNA stability of PLK1. Alternatively, PAIP1 may form a complex with PLK1 to affect cell cycle progression. Testing these hypotheses is a worthwhile endeavor and may provide insights into the clinical treatment of gallbladder carcinogenesis.
Acknowledgments
The authors are grateful to the donors who participated in this program.
Funding: This work was supported by The Natural Science Foundation of Hubei Province of China (No. 2019CFB720 to YX), the Chinese Society of Clinical Oncology (No. Y-MX2016-051 to HM), and the Application Fundamental Frontier Foundation of Wuhan (No. 2020020601012225 to HD).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-2417
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-2417
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-2417) The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All animal experiments conformed to the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health. The experiments were approved by the Ethical Committee of the Tongji Medical College, Huazhong University of Science and Technology (No. GSGC0162883). All procedures performed in this study involving human participants were performed in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Tongji Medical College, Huazhong University of Science and Technology (No. LLHBCH2019LW-003), and informed consent was provided by all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Apodaca-Rueda M, Cazzo E, De-Carvalho RB, et al. Prevalence of gallbladder cancer In patients submitted to cholecystectomy: experience of the University Hospital, Faculty of Medical Sciences, State University of Campinas - UNICAMP. Rev Col Bras Cir 2017;44:252-6. [Crossref] [PubMed]
- Chen C, Geng Z, Shen H, et al. Long-Term Outcomes and Prognostic Factors in Advanced Gallbladder Cancer: Focus on the Advanced T Stage. PLoS One 2016;11:e0166361 [Crossref] [PubMed]
- Schmidt MA, Marcano-Bonilla L, Roberts LR. Gallbladder cancer: epidemiology and genetic risk associations. Chin Clin Oncol 2019;8:31. [Crossref] [PubMed]
- Valle J, Wasan H, Palmer DH, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med 2010;362:1273-81. [Crossref] [PubMed]
- Roy G, De Crescenzo G, Khaleghpour K, et al. Paip1 interacts with poly(A) binding protein through two independent binding motifs. Mol Cell Biol 2002;22:3769-82. [Crossref] [PubMed]
- Gray NK, Coller JM, Dickson KS, et al. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J 2000;19:4723-33. [Crossref] [PubMed]
- Martineau Y, Derry MC, Wang X, et al. Poly(A)-binding protein-interacting protein 1 binds to eukaryotic translation initiation factor 3 to stimulate translation. Mol Cell Biol 2008;28:6658-67. [Crossref] [PubMed]
- Martineau Y, Wang X, Alain T, et al. Control of Paip1-eukayrotic translation initiation factor 3 interaction by amino acids through S6 kinase. Mol Cell Biol 2014;34:1046-53. [Crossref] [PubMed]
- He Y, Lin Y, Zhu Y, et al. Murine PAIP1 stimulates translation of spermiogenic mRNAs stored by YBX2 via its interaction with YBX2†. Biol Reprod 2019;100:561-72. [Crossref] [PubMed]
- Ivanov A, Shuvalova E, Egorova T, et al. Polyadenylate-binding protein-interacting proteins PAIP1 and PAIP2 affect translation termination. J Biol Chem 2019;294:8630-9. [Crossref] [PubMed]
- Grosset C, Chen CY, Xu N, et al. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 2000;103:29-40. [Crossref] [PubMed]
- Guan H, Li N, Wang X, et al. Role of Paip1 on angiogenesis and invasion in pancreatic cancer. Exp Cell Res 2019;376:198-209. [Crossref] [PubMed]
- Wang Y, Piao J, Wang Q, et al. Paip1 predicts poor prognosis and promotes tumor progression through AKT/GSK-3β pathway in lung adenocarcinoma. Hum Pathol 2019;86:233-42. [Crossref] [PubMed]
- Wang Q, Han A, Chen L, et al. Paip1 overexpression is involved in the progression of gastric cancer and predicts shorter survival of diagnosed patients. Onco Targets Ther 2019;12:6565-76. [Crossref] [PubMed]
- Piao J, Chen L, Jin T, et al. Paip1 affects breast cancer cell growth and represents a novel prognostic biomarker. Hum Pathol 2018;73:33-40. [Crossref] [PubMed]
- Kim H, Jung W, Kim A, et al. High Paip1 Expression as a Potential Prognostic Marker in Hepatocellular Carcinoma. In Vivo 2020;34:2491-7. [Crossref] [PubMed]
- Zhao MJ, Xie J, Shu WJ, et al. MiR-15b and miR-322 inhibit SETD3 expression to repress muscle cell differentiation. Cell Death Dis 2019;10:183. [Crossref] [PubMed]
- Cheng X, Hao Y, Shu W, et al. Cell cycle-dependent degradation of the methyltransferase SETD3 attenuates cell proliferation and liver tumorigenesis. J Biol Chem 2017;292:9022-33. [Crossref] [PubMed]
- Goeppert B, Truckenmueller F, Ori A, et al. Profiling of gallbladder carcinoma reveals distinct miRNA profiles and activation of STAT1 by the tumor suppressive miRNA-145-5p. Sci Rep 2019;9:4796. [Crossref] [PubMed]
- Pang K, Hao L, Shi Z, et al. Comprehensive gene expression analysis after ERH gene knockdown in human bladder cancer T24 cell lines. Gene 2020;738:144475 [Crossref] [PubMed]
- Zhang J, Wang Y, Shen Y, et al. G9a stimulates CRC growth by inducing p53 Lys373 dimethylation-dependent activation of Plk1. Theranostics 2018;8:2884-95. [Crossref] [PubMed]
- Larochelle M, Bergeron D, Arcand B, et al. Proximity-dependent biotinylation mediated by TurboID to identify protein-protein interaction networks in yeast. J Cell Sci 2019;132:jcs232249 [Crossref] [PubMed]
- Personeni N, Lleo A, Pressiani T, et al. Biliary Tract Cancers: Molecular Heterogeneity and New Treatment Options. Cancers (Basel) 2020;12:3370. [Crossref] [PubMed]
- Crowley LC, Marfell BJ, Scott AP, et al. Quantitation of Apoptosis and Necrosis by Annexin V Binding, Propidium Iodide Uptake, and Flow Cytometry. Cold Spring Harb Protoc 2016; [Crossref] [PubMed]
- Degterev A, Boyce M, Yuan J. A decade of caspases. Oncogene 2003;22:8543-67. [Crossref] [PubMed]
- O'Brien MA, Daily WJ, Hesselberth PE, et al. Homogeneous, bioluminescent protease assays: caspase-3 as a model. J Biomol Screen 2005;10:137-48. [Crossref] [PubMed]
- Bruinsma W, Raaijmakers JA, Medema RH. Switching Polo-like kinase-1 on and off in time and space. Trends Biochem Sci 2012;37:534-42. [Crossref] [PubMed]
- Li W, Wang HY, Zhao X, et al. A methylation-phosphorylation switch determines Plk1 kinase activity and function in DNA damage repair. Sci Adv 2019;5:eaau7566 [Crossref] [PubMed]
- Gutteridge RE, Ndiaye MA, Liu X, et al. Plk1 Inhibitors in Cancer Therapy: From Laboratory to Clinics. Mol Cancer Ther 2016;15:1427-35. [Crossref] [PubMed]
- Wang R, Song Y, Xu X, et al. The expression of Nek7, FoxM1, and Plk1 in gallbladder cancer and their relationships to clinicopathologic features and survival. Clin Transl Oncol 2013;15:626-32. [Crossref] [PubMed]
- Valle JW, Lamarca A, Goyal L, et al. New Horizons for Precision Medicine in Biliary Tract Cancers. Cancer Discov 2017;7:943-62. [Crossref] [PubMed]
- Hezel AF, Deshpande V, Zhu AX. Genetics of biliary tract cancers and emerging targeted therapies. J Clin Oncol 2010;28:3531-40. [Crossref] [PubMed]
(English Language Editor: J. Gray)


