
LncRNA TCONS_00004099-derived microRNA regulates oncogenesis through PTPRF in gliomas
Introduction
Glioma derived from the neural ectoderm is the most common type of primary malignant brain cancer and accounting for 50% of primary brain malignancies(1).The poor prognosis in patients with glioma is due in part to the strong tumor growth capacity of cancer cells and the tumor's resistance to radiotherapy and chemotherapy induced apoptosis (2). As a chemotherapeutic agent, temozolomide (TMZ) is a first-line therapy for gliomas, especially in glioblastoma multiforme (GBM), and has achieved remarkable success (3). However, a large proportion of patients develop tumor resistance to TMZ due to the uncontrolled proliferation of glioma cells and tumor growth (4). Therefore, in addition to understanding the normal oncological behaviors of gliomas (cell viability, proliferation, invasion, apoptosis, and cell cycle), it is also important to explore the manifestations of chemoresistance.
Long non-coding RNA (lncRNA) is a section of non-coding RNA greater than 200 nucleotides with no potential to translate into protein (5). The functions of lncRNAs have been associated with microRNAs (miRNAs) and have been shown to be involved in gene regulation of gliomas (6). As a regulatory molecule, lncRNAs have diverse functions in cell cycle, proliferation, invasion and survival (7,8). Moreover, it was shown that lncRNAs in gliomas has high rates of chemoresistance and recurrence (9-11). Therefore, revealing the mechanisms that mediate the chemoresistance of gliomas is urgently needed. Analysis of sequencing data identified a new lncRNA, named lncRNA TCONS_00004099, which could derive a new microRNA and was highly expressed in glioma. The sequence of the new miRNA is “UGUGGUUCUGGGGCCAUUGUU”. Bioinformatics analysis shows that lncRNA TCONS_00004099 can target protein mRNA depending on the new microRNA which derives from such lncRNA via miRNA binding site prediction software miRanda. 21 nucleotides generated by lncRNA TCONS_00004099 splicing is predicted to bind to PTPRF mRNA and inhibit the translation.
miRNAs are non-coding single-stranded RNA with 18 to 24 nucleotides. miRNAs inhibit the translation of target mRNAs by binding to the 3'-untranslated region (3' UTR) of different target mRNAs (12). In recent years, hundreds of different miRNAs have been identified in cancers. For example, the STAT3/miR-182-5p/PCDH8 axis has been shown to play a key role in glioma tumorigenesis (13). However, the role of miRNAs derived from lncRNA in glioma oncogenesis is still not clear.
Protein tyrosine phosphatase (PTP) works with protein tyrosine kinases to regulate many important signaling pathways in cancer (14). The PTP family constructed of 107 members which divided into 4 subfamilies. PTPRF belongs to class I PTP and is closely related to PTPRD and PTPRS (15). Previous research has shown that MicroRNA-24 enhances breast cancer invasion and metastasis by targeting PTPN9 and PTPRF to promote EGF signaling (16). However, a systematic understanding of how lncRNA contributes to PTPRF in gliomas is lacking. Hence, there is an urgent need to identify novel PTPRF-related lncRNA and investigate their potential mechanisms of oncogenesis in gliomas.
In this study, we investigated the role and the mechanism of lncRNA TCONS_00004099 in the oncogenesis of gliomas. Moreover, we determined whether lncRNA TCONS_00004099 regulated the migration, invasion, tumor growth, and TMZ resistance of glioma through generating microRNAs. Importantly, we determined that lncRNA TCONS_00004099 promotes the oncogenesis of glioma by generating microRNA and targeting PTPRF.
We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-2442).
Methods
Clinical samples and cell lines
Human glioma samples were obtained from the Neurosurgery Department of the Sun Yat-sen Memorial Hospital, Sun Yat-sen University. Different grades of glioma tissues were obtained from different glioma patients. Normal brain tissues were obtained from patients who required partial brain excision such as patients with traumatic brain injury. All tissues were preserved in liquid nitrogen for subsequent total RNA extraction. The study was approved by institutional ethics board of Sun Yat-sen Memorial Hospital, Sun Yat-sen University (No.: 2019060) and conformed to the guidelines of the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from the patients before the start of the study. The U87 and U251 glioma cell lines, and the HEB cell line were purchased from Shanghai Institutes for Biological Sciences of Chinese Academy of Sciences (SIBS, Shanghai) and cultured in Dulbecco modified Eagle medium (DMEM, GIBCO) supplemented 10% fetal bovine serum (FBS, GIBCO). The glioma cells were incubated in 37 °C in humid atmosphere with 5% CO2.
Oligonucleotide synthesis and transfection, lentivirus vector transfection
Cells were transfected with lentivirus vector pZHLV-CMV-zsGreen1-puro packaged with the lncRNA TCONS_00004099 overexpression sequence or short hairpin RNA (shRNA). The synthetic inhibitor or mimics of miRNA derived from lncRNA TCONS_00004099, and small interfering RNA (siRNA) were transfected into cell lines using Lipofectamine 3000 (Thermo, MA, USA). All the oligonucleotides and scramble sequences were designed and provided by GenePharma (Shanghai, China). Lentivirus vector pZHLV-CMV-zsGreen1-puro packaged by DongBio.Co.Ltd (Shenzhen, China). The sequences of lncRNA TCONS_00004099 and the related miRNA were demonstrated in Supplementary file.
RNA extraction and qPCR
RNA was isolated from tissues and cells using the Trizol Reagent (TAKARA, Japan). The total RNA was reverse transcribed into cDNA using the Master Mix kit (TAKARA, Japan). Then, the Mir-X miRNA First-Strand Synthesis Kit (TAKARA, Japan) was used in the transcription of miRNA. Quantitative Real-time PCR (qPCR) was carried out with SYBR-Green Master Kit (TAKARA, Japan). The expression levels of target genes were normalized to glyceraldehyde 3-phosphate dehydrogenase (GADPH). The forward primer sequence of LncRNA TCONS_00004099 was 5'-CCTAACAGGTGCTGTGGCA-3', while the reverse sequence was 5'-TCATCGTTGCAGAACAATGGC -3'. The forward primer sequence of GAPDH was 5'-GCTCTCTGCTCCTCCTGTTC-3', while the reverse primer sequence was 5'-AAATGAGCCCCAGCCTTCTC-3'.
Cell Counting Kit 8 (CCK8) assay
Cells were harvested and inoculated into 96-well plate (3,000–6,000 cells/per well) and cultured for 24 hours. After treatment, the culture medium was mixed with the CCK8 solution (the proportion was 1:10) and added to each well. Cells were incubated at 37 °C for 1–2 hours. Thereafter, the absorbance of each well was measured at a wavelength of 450 nm.
Wound healing assay and transwell test
Glioma cells were cultured in six-well plates under different treatment conditions. After 72 hours, the culture media was removed, serum-free medium was added to treat the cells for another 12 hours. A straight wound was scratched using a 200 µL-pipette tip. The cells would migrate through the scratch and heal the wound. Photographs were taken using an inverted microscope at 0, 12, and 24 hours.
To further detect the migration and invasion ability, transwell assays were performed using transwell inserts (Costar, USA) coated with 50 µL Matrigel Matrix (BD, USA). The inserts were put into 24-well plates. Cells in FBS-free DMEM were seeded into the upper chambers while the lower chambers were filled with DMEM with 10% FBS. After 24 hours, the excess cells were wiped off with a cotton swab. 4% paraformaldehyde were used to fixed chambers and then stained with 0.1% crystal violet dye. Cells were counted with an inverted microscope (Olympus).
Colony formation assays
Glioma cells were cultured in six-well plates under different treatment conditions for 48 hours. 4% paraformaldehyde were used to fixed cells and then stained with 0.1% crystal violet dye. The number of colonies per well was counted with an inverted microscope (Olympus).
Apoptosis applications
Cells were cultured and treated in combination with 300 µM temozolomide (TMZ). After 72 hours, cells were digested, centrifuged, and washed with PBS. Binding buffer (200 mL) and Annexin V (5 µL) from the Annexin V/PI Kit (BD, USA) were added to the cells and incubated in the dark for 15 minutes. Thereafter, 10 mL propidium iodide (PI) was added and flow cytometry was performed immediately at 488 and 594 nm.
Western blot analysis
Protein concentrations were determined using the G250 assay (Keygen). Proteins were separated using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) by electrophoresis, then transferred to polyvinylidene difluoride (PVDF) membranes. A PTPRF monoclonal antibody (rabbit; 1:2,000; Abcam, UK) were incubated with the PVDF membranes overnight, followed by incubation with an anti-rabbit secondary antibody (1:10,000; Proteintech, USA). The immune complexes were detected using the Two-color Infrared Laser Imaging System (Odyssey).
Zebrafish xenograft model
The Zebrafish xenograft model is a one-way trial that transgenic A/B zebrafish line (Zebrafish Technology Platform, Sun Yat-sen University) was used in this study. Zebrafish embryos were raised, staged, and maintained according to standard procedures. Zebrafish embryos (no gender distinction) were anesthetized with 0.02% tricaine (Sigma) and positioned on a 10-cm Petri dish coated with 3% agarose. Transfected cells (approximately 300 cells) were injected into the yolk sac of zebrafish embryos 2 days-post fertilization using a Picospritzer (Parker Inc.). After 24 hours, zebrafish were included in the study if the fluorescence foci in the yolk sacs was similar. If any zebrafish had less fluorescence or no fluorescence foci than the other groups, they were not included in the study. After implantation with transfected glioma cells, zebrafish embryos (including non-implanted controls) were maintained at 33 °C for the next 96 hours. 10 embryos were included for each cell line or condition and a total of 60 embryos were included in each experiment, zebrafish embryos were placed in 96-well plates and randomly divided into each treatment group using a randomization table (http://www.graphpad.com/quickcalcs/randomN2/). For each zebrafish, three different investigators were involved as follows: a first investigator administered the transfected cell. This investigator was the only person aware of the treatment group allocation. A second investigator was responsible for the injected procedure, whereas a third investigator monitored the fluoresce in the yolk sac.
A protocol was prepared before the study without registration. Experiments were performed under a project license (No.: LL-KY-2019105) granted by Institute Ethics Committee, Shenzhen People’s Hospital, in compliance with China national or institutional guidelines for the care and use of animals.
Statistical analysis
For unpaired samples, Student’s t-test was used to analyze statistical differences. For multiple comparisons with one control group, an analysis of variance followed by the Dunnett test was performed. A P value less than 0.05 was considered statistically significant. All results were independently repeated at least three times.
Results
LncRNA TCONS_00004099 was upregulated in glioma tissues and cell lines, while knock down LncRNA TCONS_00004099 downregulated glioma cells viability, invasion, and migration in vitro
The expression levels of lncRNA TCONS_00004099 in different GBM tissues were assessed using qPCR. The levels of lncRNA TCONS_00004099 were markedly upregulated in GBM tissues compared to normal brain tissues. In addition, lncRNA TCONS_00004099 expression was remarkable higher in the U87 and U251 glioma cell lines compared to the HEB cell line (Figure 1A). These results suggested that lncRNA TCONS_00004099 is positively correlated with human glioma.
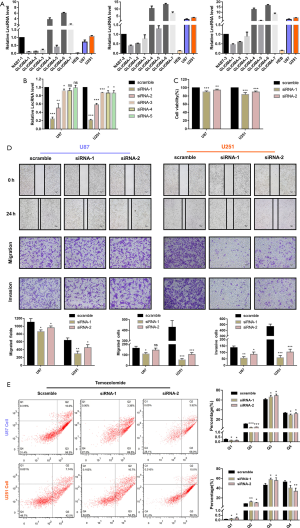
To knock down lncRNA TCONS_00004099, five siRNAs that interfered with the lncRNA in U87 and U251 cells were assessed. The qPCR results showed that siRNA1 and siRNA2 downregulated the levels of lncRNA TCONS_00004099 most significantly (Figure 1B). The sequence of siRNA1 and siRNA2 is selected for further experiment. CCK8 assays showed that cellular viability was suppressed markedly by siRNA1 and siRNA2 (Figure 1C). Transwell migration tests, transwell invasion assays, and wound healing assays showed that siRNA1 and siRNA2 inhibited the migration and invasion of U87 and U251 cells (Figure 1D). Furthermore, 300µM temozolomide (TMZ) was used to generate an apoptosis model, and the results demonstrated that knockdown of lncRNA TCONS_00004099 significantly enhanced apoptosis in U87 and U251 cells compared to control cells (Figure 1E).
These data suggested that lncRNA TCONS_00004099 expression was increased in glioma tissues and glioma cell lines. Downregulation of lncRNA TCONS_00004099 significantly decreased cell growth, migration, and invasion, and enhanced TMZ-induced apoptosis.
LncRNA TCONS_00004099 enhanced glioma cell viability and proliferation through generating miRNA TCONS_00004099 and targeting PTPRF
Glioma cell lines were transfected with the overexpression lentivirus vector or shRNA lentivirus vector, which upregulated or downregulated the levels of lncRNA TCONS_00004099, respectively. The upregulated-lncRNA group was named as the overexpression group (OE) while the downregulated-lncRNA group was named as the low-expression group (LE). The expression levels in each group were confirmed by qPCR assay (Figure 2A).

Specific qPCR was performed to identify cellular miRNAs and a new miRNA derived from lncRNA TCONS_00004099 named miRNA TCONS_00004099 was identified. The miRNA inhibitor or mimics was transfected into cells to reverse the effects of the lncRNA. The 7 experimental groups included scramble sequences group(scramble), miRNA inhibitor group (miR-IH), miRNA mimics group (miR-MI), lncRNA overexpression group (OE), lncRNA expression plus miRNA inhibitor group (OE + IH), lncRNA low-expression group (LE), and lncRNA low-expression plus miRNA mimics group (LE + MI). Assessing the miRNA TCONS_00004099 levels revealed that both lncRNA TCONS_00004099 and the miRNA mimics increased the levels of cellular miRNA TCONS_00004099 (Figure 2B).
CCK8 assays demonstrated that the effects of miRNA TCONS_00004099 on cell viability corresponded to that of lncRNA TCONS_00004099. Overexpression of the lncRNA significantly promoted cell growth and this was reversed by the miRNA inhibitor. Downregulation of lncRNA suppressed cell growth and this was reversed by the miRNA mimics (Figure 2C). Colony formation assays revealed similar results on cell proliferation in both U87 and U251 cell lines (Figure 2D).
Wound healing assays, transwell migration assays, and transwell invasion assays also demonstrated that overexpression of lncRNA TCONS_00004099 enhanced migration and invasion, while knockdown lncRNA decreased the capacity of migration and invasion of U87 and U251 cell. These effects were reversed by both the miRNA inhibitor and the miRNA mimics (Figure 3A,B).
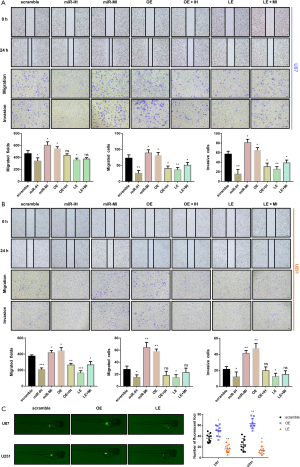
Glioma Cells in the OE and LE group were injected into the zebrafish tumor model and the invasive ability was assessed by counting the number of fluorescent foci at 96 hours after injection. The results were similar to that observed in vitro (Figure 3C).
Moreover, Flow cytometry assays revealed that lncRNA TCONS_00004099 significantly decreased cell apoptosis and enhanced cell proliferation. In contrast, down-regulation of lncRNA TCONS_00004099 decreased proliferation and promoted cell apoptosis in U87 and U251 cells (Figure 4A,B).
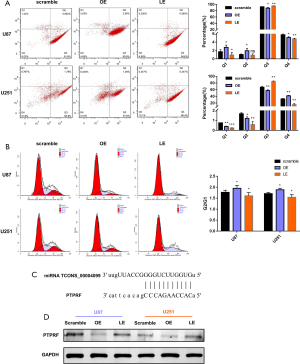
Then, we analyzed potential targets of miRNA TCONS_00004099.There were a great number of candidates potentially targeted by miRNA TCONS_00004099, then we finally focused on PTPRF. The potential binding sites appeared nucleotides 6827 to 6837 for PTPRF mRNA (NM_002840) (Figure 4C). Meanwhile, Western blot analyses were used to detect the expression of the miRNA TCONS_00004099-targeted PTPRF in U87 and U251 cells. Overexpression of lncRNA TCONS_00004099 suppressed PTPRF expression in U87 and U251 cells, while knockdown of lncRNA TCONS_00004099 increased PTPRF expression in U87 and U251 cells (Figure 4D). These results showed that LncRNA TCONS_00004099 enhanced the viability and proliferation through generating miRNA TCONS_00004099 and targeting PTPRF in glioma cells.
Discussion
Nervous system neoplasms account for about 3% of all cancers (17,18), with a mortality rate of 3.4 per 100,000 people (19). Numerous studies have identified non-coding RNAs (ncRNAs) as novel markers in carcinogenesis, among which, lncRNAs appear to be the most promising. Although the most well-known and examined ncRNAs are the miRNAs and it is frequently disrupted in brain tumors (20). However, lncRNA-derived miRNA is much more stable and effective in gliomas, with lncRNAs acting as a repository that is hard to degrade. The function of miRNAs and its interaction with lncRNA in brain tumors have been summarized by different reports (21). miRNAs and lncRNAs were key regulators that mediate tumor growth and oncological behaviors, including tumor proliferation, invasion, apoptosis, and cell metabolism (22). Indeed, lncRNAs have been associated with tumor progression, apoptosis, and cell cycle in gliomas (23). For instance, lncRNA RAMP2-AS1 is remarkably downregulated in GBM and is involved in regulating the cell cycle (24), while lncRNA FOXD2-AS1 promotes glioma progression by regulating cyclin-dependent kinases (25).
Our study shown that the expression of lncRNA TCONS_00004099 was markedly higher in glioma tissues and cell lines compared to normal human brain tissues and normal HEB cell lines. Silencing lncRNA TCONS_00004099 with siRNA limited glioma cell viability, migration, and invasion in U87 and U251 cells, and enhanced apoptosis. Considering the roles of these small non-coding RNAs, novel therapeutic strategies involving miRNA silencing or miRNA mimics may be considered in glioma management.
Our investigation further identified a new miRNA derived from lncRNA TCONS_00004099 and revealed that miRNA TCONS_00004099 increased cell viability and reduced apoptosis in U87 and U251 cells, thereby promoting glioma growth. The miRNA inhibitor significantly suppressed cell migration and invasion, and reversed the effects of lncRNA TCONS_00004099.
Currently, the most common treatment for glioma is maximal surgical resection. Post-surgery radiotherapy and chemotherapy with temozolomide (TMZ) are also first-line treatments for gliomas resulting in higher survival rates in patients (26). However, recurrence and tumor resistance to TMZ is not uncommon and can lead to treatment failure. In this study, cells were treated with the combination therapy of TMZ (300 µM) and downregulation of lncRNA TCONS_00004099. Knockdown of such lncRNA, as well as the miRNA inhibitor both significantly enhanced the TMZ-induced apoptosis compared to the control group. The effects suggested that lncRNA TCONS_00004099 and its associated miRNA may be potential therapeutic targets in combination with TMZ.
Protein Tyrosine Phosphatase Receptor Type F (PTPRF) has a complex role in the regulatory function of cancers. The role of PTPRF in cancer is contradictory, suggesting that PTPRF acts either as an oncogene or as a tumor suppressor in different types of cancer (16,27-32). Recent reports have demonstrated that increased levels of PTPRF are associated with poor prognosis in colorectal cancer patients (14), but the expression and the role of PTPRF and in gliomas still unknown.
Although the relationship between miRNA and PTPRF has been studied (16), but the relationship between PTPRF and lncRNA has so far remained unclear. Thus, in our study, western blot analyses showed that lncRNA TCONS_00004099 was enriched in glioma cells and its overexpression was associated with lower PTPRF expression in both U87 and U251 cells. Interestingly, decreased expression of lncRNA TCONS_00004099 resulted in higher PTPRF expression in U87 and U251 cells, which may represent the complex regulatory role of PTPRF. Nonetheless, xenograft zebrafish models and cell function experiments demonstrated that inhibiting lncRNA TCONS_00004099 blocked tumor proliferation and invasion both in vitro and in vivo. In fact, zebrafish share up to 85% homology with human genes, and its signal pathway is basically similar to that of human beings. Its biological structure and physiological functions are highly similar to those of mammals and the microenvironment in zebrafish embryo is purer which is suitable for study tumor proliferation and invasion (33). However, the influence of zebrafish gender on the results was not studied in our study, which is also needed to be supplemented in subsequent experiments. Many studies have demonstrated the competitive endogenous RNA (ceRNA) network where lncRNAs act as miRNA sponges to regulate mRNA expression in tumor development. This study, however, described a different type of interaction between lncRNA and miRNA, lncRNA could generate miRNA by RNA splicing and work through miRNA regulation mechanism. miRNA TCONS_00004099 targeted PTPRF mRNA and combine with complementary sequences of PTPRF mRNA, inhibit mRNA translation and promote mRNA degradation. The lncRNA TCONS_00004099 promoted cell proliferation and invasion through the derived miRNA (miRNA TCONS_00004099) and its target PTPRF. Significantly, the lncRNA TCONS_00004099/miRNA TCONS_00004099/PTPRF axis may be a critical target for glioma diagnosis and treatment.
Acknowledgments
Funding: This work was supported by grants from the Guangdong Natural Science Foundation (No. 2020A1515010289); Shenzhen People’s Hospital Young and Middle-aged Research Funds Cultivation Fund (No. SYKYPY201923); Shenzhen Key Medical Discipline Construction Fund (Shenzhen Key Medical Discipline 2020-2024); National Natural Science Foundation of China (Award Number: 81602205) and the Young Teacher Training Project of Sun Yat-sen University (Award Number: 20ykpy97).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-2442
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-2442
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-2442). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by institutional ethics board of Sun Yat-sen Memorial Hospital, Sun Yat-sen University (No.: 2019060) and conformed to the guidelines of the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from the patients before the start of the study. Experiments were performed under a project license (No.: LL-KY-2019105) granted by Institute Ethics Committee, Shenzhen People’s Hospital, in compliance with China national or institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ostrom QT, Bauchet L, Davis FG, et al. The epidemiology of glioma in adults: a "state of the science" review. Neuro Oncol 2014;16:896-913. [Crossref] [PubMed]
- Krakstad C, Chekenya M. Survival signalling and apoptosis resistance in glioblastomas: opportunities for targeted therapeutics. Mol Cancer 2010;9:135. [Crossref] [PubMed]
- Jiapaer S, Furuta T, Tanaka S, et al. Potential Strategies Overcoming the Temozolomide Resistance for Glioblastoma. Neurol Med Chir (Tokyo) 2018;58:405-21. [Crossref] [PubMed]
- Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell 2012;10:717-28. [Crossref] [PubMed]
- Castro-Oropeza R, Melendez-Zajgla J, Maldonado V, et al. The emerging role of lncRNAs in the regulation of cancer stem cells. Cell Oncol (Dordr) 2018;41:585-603. [Crossref] [PubMed]
- Liao Y, Shen L, Zhao H, et al. LncRNA CASC2 Interacts With miR-181a to Modulate Glioma Growth and Resistance to TMZ Through PTEN Pathway. J Cell Biochem 2017;118:1889-99. [Crossref] [PubMed]
- Fu PF, Zheng X, Fan X, et al. Role of cytoplasmic lncRNAs in regulating cancer signaling pathways. J Zhejiang Univ Sci B 2019;20:1-8. [Crossref] [PubMed]
- Guo JC, Yang YJ, Zheng JF, et al. Silencing of long noncoding RNA HOXA11-AS inhibits the Wnt signaling pathway via the upregulation of HOXA11 and thereby inhibits the proliferation, invasion, and self-renewal of hepatocellular carcinoma stem cells. Exp Mol Med 2019;51:1-20. [Crossref] [PubMed]
- Bi CL, Liu JF, Zhang MY, et al. LncRNA NEAT1 promotes malignant phenotypes and TMZ resistance in glioblastoma stem cells by regulating let-7g-5p/MAP3K1 axis. Biosci Rep 2020;40:BSR20201111 [Crossref] [PubMed]
- Tang G, Luo L, Zhang J, et al. lncRNA LINC01057 promotes mesenchymal differentiation by activating NF-κB signaling in glioblastoma. Cancer Lett 2021;498:152-64. [Crossref] [PubMed]
- Mu Y, Tang Q, Feng H, et al. lncRNA KTN1-AS1 promotes glioma cell proliferation and invasion by negatively regulating miR-505-3p. Oncol Rep 2020;44:2645-55. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Xue J, Zhou A, Wu Y, et al. miR-182-5p Induced by STAT3 Activation Promotes Glioma Tumorigenesis. Cancer Res 2016;76:4293-304. [Crossref] [PubMed]
- Gan T, Stevens AT, Xiong X, et al. Inhibition of protein tyrosine phosphatase receptor type F suppresses Wnt signaling in colorectal cancer. Oncogene 2020;39:6789-801. [Crossref] [PubMed]
- Alonso A, Sasin J, Bottini N, et al. Protein tyrosine phosphatases in the human genome. Cell 2004;117:699-711. [Crossref] [PubMed]
- Du WW, Fang L, Li M, et al. MicroRNA miR-24 enhances tumor invasion and metastasis by targeting PTPN9 and PTPRF to promote EGF signaling. J Cell Sci 2013;126:1440-53. [Crossref] [PubMed]
- Mortazavi SMJ, Mortazavi SAR, Paknahad M. Cancers of the Brain and CNS: Global Patterns and Trends in Incidence. J Biomed Phys Eng 2018;8:151-2. [PubMed]
- Miranda-Filho A, Piñeros M, Soerjomataram I, et al. Cancers of the brain and CNS: global patterns and trends in incidence. Neuro Oncol 2017;19:270-80. [PubMed]
- . GBD 2016 Brain and Other CNS Cancer Collaborators. Global, regional, and national burden of brain and other CNS cancer, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol 2019;18:376-93. [PubMed]
- Turner JD, Williamson R, Almefty KK, et al. The many roles of microRNAs in brain tumor biology. Neurosurg Focus 2010;28:E3 [Crossref] [PubMed]
- Nicoloso MS, Calin GA. MicroRNA involvement in brain tumors: from bench to bedside. Brain Pathol 2008;18:122-9. [Crossref] [PubMed]
- Gournay M, Paineau M, Archambeau J, et al. Regulat-INGs in tumors and diseases: Focus on ncRNAs. Cancer Lett 2019;447:66-74. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100:57-70. [Crossref] [PubMed]
- Liu S, Mitra R, Zhao MM, et al. The Potential Roles of Long Noncoding RNAs (lncRNA) in Glioblastoma Development. Mol Cancer Ther 2016;15:2977-86. [Crossref] [PubMed]
- Wang J, Li B, Wang C, et al. Long noncoding RNA FOXD2-AS1 promotes glioma cell cycle progression and proliferation through the FOXD2-AS1/miR-31/CDK1 pathway. J Cell Biochem 2019;120:19784-95. [Crossref] [PubMed]
- Weller M, van den Bent M, Tonn JC, et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol 2017;18:e315-29. [Crossref] [PubMed]
- Wang X, Liu T, Bai Y, et al. Polymerase I and transcript release factor acts as an essential modulator of glioblastoma chemoresistance. PLoS One 2014;9:e93439 [Crossref] [PubMed]
- Nunes-Xavier CE, Martín-Pérez J, Elson A, et al. Protein tyrosine phosphatases as novel targets in breast cancer therapy. Biochim Biophys Acta 2013;1836:211-26. [PubMed]
- Levea CM, Mcgary CT, Symons JR, et al. PTP LAR expression compared to prognostic indices in metastatic and non-metastatic breast cancer. Breast Cancer Res Treat 2000;64:221-8. [Crossref] [PubMed]
- Wang Z, Shen D, Parsons DW, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science 2004;304:1164-6. [Crossref] [PubMed]
- Bera R, Chiou CY, Yu MC, et al. Functional genomics identified a novel protein tyrosine phosphatase receptor type F-mediated growth inhibition in hepatocarcinogenesis. Hepatology 2014;59:2238-50. [Crossref] [PubMed]
- DaSilva JO, Amorino GP, Casarez EV, et al. Neuroendocrine-derived peptides promote prostate cancer cell survival through activation of IGF-1R signaling. Prostate 2013;73:801-12. [Crossref] [PubMed]
- Osmani N, Goetz JG. Multiscale Imaging of Metastasis in Zebrafish. Trends Cancer 2019;5:766-78. [Crossref] [PubMed]


