
Long non-coding RNA MEG3 regulates the progress of osteoarthritis by regulating the miR-34a/Klotho axis
Introduction
Osteoarthritis (OA) is a chronically progressive disease that is characterized by the chronic degradation of articular cartilage, subchondral bone sclerosis, and joint margin degeneration (1). Articular cartilage is a highly differentiated organic tissue composed of a small amount of chondrocytes and a large amount of extracellular matrix, which can resist external mechanical pressure (2). At present, OA is generally believed to be caused by a variety of factors, such as biomechanics, genetics, metabolic distress, chondrocyte death, and cytokines. An epidemiological survey of a large number of people found that the abnormal biomechanical factors of joints dominate the pathogenesis of OA (3). The main symptom of OA is joint pain and limited mobility with gradual development. This symptom affects the patient’s daily life and can encroach upon their ability to work. Current methods for treating OA are mainly divided into nonsurgical and surgical treatment methods. Nonsurgical treatment focuses on pain management, and the most common method of surgical treatment is artificial joint replacement (1,4). Although artificial joint replacement can relieve the patient’s symptoms and improve their quality of life, there are deficiencies associated with this approach. For example, its high cost exerts severe pressure on the patient’s family and social medical resources.
Non-coding RNA is an important regulator of inflammation and immune response (5). Long non-coding RNA (lncRNA) is non-coding RNA with a length of more than 200 nucleotides and highly tissue-specific expression (6). It is involved in many biological processes, such as homeostasis maintenance in cells and tissues (7). In the immune system, lncRNA can regulate the differentiation, activation, proliferation, apoptosis, and cytokine expression of T lymphocytes (8,9). The abnormal expression of lncRNA has been observed in various tumors and diseases (10). Some lncRNAs can be used as diagnostic or prognostic markers and potential therapeutic targets. In addition, they are considered endogenous microRNA (miRNA) sponges (11). The lncRNA maternally expressed gene 3 (MEG3) is an imprinted gene that is located on human chromosome 14q32.3 (12). It can reduce high glucose-induced inflammation and the apoptosis of retinal epithelial cells by regulating the miR-34a/SIRT1 axis (13). During liver ischemia-reperfusion, lncRNA MEG3 protects liver cells from ischemia-reperfusion injury by down-regulating the expression of miR-34a (14). This indicates that lncRNA MEG3 also plays an important role in the pathogenesis of inflammation, and OA is also a common inflammatory disease. Therefore, we explored the specific mechanism of lncRNA MEG3 in the course of OA.
The miRNAs are a type of small non-coding RNAs with lengths of 21–23 nucleotides; they also play an important regulatory role in various biological processes (15). They can inhibit protein expression or induce messenger RNA (mRNA) degradation by undergoing complementary pairing with bases in the specific site of the target gene mRNA to participate in the regulation of gene expression (16). The tumor suppressor gene p53 can directly induce miR-34 expression, thereby triggering cell apoptosis and cell cycle arrest (17,18). Moreover, studies have also shown that Mir-34A is elevated in synovial fluid of patients with advanced knee arthritis (19). The lncRNA MEG3 and miR-34a have 3 binding sites (20). The loss of cartilage in vivo caused by chondrocyte death is significantly suppressed after knocking out miR-34a (20). The gene Klotho is closely related to human aging, and its role in maintaining kidney function and preventing kidney failure is currently attracting great attention (21). The extrarenal expression of Klotho is limited to a few tissues, such as skeletal muscle and the placenta. Klotho is remarkably expressed in osteoblasts (22). Klotho gene-deficient mice exhibit a series of phenotypic changes similar to human aging; these changes include growth arrest, arteriosclerosis, osteoporosis, and metabolic dysfunction (23). The Klotho protein can exert antioxidative stress by inhibiting insulin growth factors and finally inhibits cell apoptosis (24). It can also inhibit endoplasmic reticulum stress-mediated cell apoptosis (25). The miRNA miR-34a can promote the progression of renal fibrosis by down-regulating the expression of Klotho (24).
Therefore, we speculated that lncRNA MEG3 may regulate the progress of OA by regulating the miR-34a/Klotho axis. In this study, an animal model of OA was established to evaluate the expression of lncRNA MEG3, miR-34a, and Klotho. Moreover, we performed cell experiments to show that lncRNA MEG3 could regulate OA by regulating the miR-34a/Klotho axis. We present the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-894/rc).
Methods
Materials, cells and animals
Cysteine (C0012), lipopolysaccharide (LPS, L8880), and papain (G8430) were purchased from Solarbio (Beijing, China). HiScript II Q RT SuperMix for quantitative polymerase chain reaction (qPCR) (+gDNA wiper) (R223-01) was purchased from Vazyme (Jiangsu, China). Horseradish peroxidase-conjugated (HRP) goat anti-mouse IgG (H+L) (ZB-2305, 1/2,000), and HRP-conjugated goat anti-rabbit IgG (H+L) (ZB-2301, 1/2,000) and mouse anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) monoclonal antibody (TA-08, 1/2,000) were obtained from ZSGB (Beijing, China). An interleukin 1β (IL-1β) enzyme-linked immunosorbent assay (ELISA) kit (MM-0181H2) was provided by Meimian (Jiangsu, China). A miRNA complementary DNA (cDNA) synthesis kit (CW2141S), miRNA purification kit (CW0627S), miRNA qPCR assay kit (CW2142S) Trizon reagent (CW0580S) and ultrapure RNA extraction kit (CW0581M) were obtained from CWBio (Beijing, China). Mouse anti-transforming growth factor-β1 (TGF-β1) monoclonal antibody (ab190503, 1/1,000), rabbit anti-Bax monoclonal antibody (ab32503, 1/5,000), rabbit anti-Caspase 3 monoclonal antibody (ab32351, 1/500), rabbit anti-Caspase 8 monoclonal antibody (ab108333, 1/5,000), rabbit anti-fibroblast growth factor 23 (FGF23) polyclonal antibody (ab98000, 1/1,000), and rabbit anti-Klotho monoclonal antibody (ab181373, 1/1,000) were purchased from Abcam (Cambridge, UK). Lipofectamine 3000 reagent (L3000015) was purchased from Invitrogen (Carlsbad, CA, USA). Polyvinylidene fluoride (PVDF) membrane (IPVH00010) was obtained from Millipore (Billerica, MA, USA). The pmir-GLO plasmid was provided by General Biosystems (Anhui, China). Rabbit anti-Bcl-2 polyclonal antibody (A11025, 1/1,000) was obtained from ABclonal (Cambridge, MA, USA); 2× SYBR Green PCR Master Mix (A4004M) was purchased from Lifeint (Fujian, China).
Human normal chondrocytes C28/I2 (BNCC339995) were purchased from BeNa Culture Collection (Beijing, China) and cultured in Dulbecco’s modified Eagle medium (DMEM; Gibco, Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Gibco, USA) at 37 ℃ and 5% CO2.
Male C57BL/6 mice (4-week-old, body weight 15–18 g) were provided by Hunan SJA Laboratory Animal Co., Ltd. [License No. SCXK(Xiang)2016-0002]. The mice were housed in an environment at 18–26 ℃ and 40–70% relative humidity with free access to food and water. A protocol was prepared before the study without registration. Animal experiments were granted by the Ethics Committee of the First Affiliated Hospital of Jinan University (No. 20190550x), in compliance with the institutional guidelines of the First Affiliated Hospital of Jinan University for the care and use of animals.
OA model establishment and animal grouping
The OA model was established in accordance with the method of Cheng et al. (26). We randomly divided 12 mice into 2 groups (n=6 per group), namely, the control and model groups. The mice in the control group did not receive any treatment. The mice in the model group were weighed and injected intraperitoneally with 0.6% pentobarbital sodium at a dose of 70 mg/kg. The hair on the right knees of the mice was removed. The right knee joints of the mice were bent at 45° and injected with a mixture containing 0.1% papain and 0.03 mol/L L-cysteine at a dose of 0.1 mL/kg. After 1 week, the mice were sacrificed through cervical dislocation. The skin around the knee joints was cut to expose the knee joints. The knee joints were taken, and the muscles at the knee joint were removed.
HE staining
In accordance with a previous report (27), articular cartilages were washed with running water for several hours; dehydrated in 70%, 80%, and 90% ethanol; immersed in a mixture of equal quantities of ethanol and xylene for 15 min; immersed in xylene twice for 15 min each time until transparent; immersed in a mixture of equal parts xylene and paraffin for 15 min; waxed in paraffin twice for 50–60 min each time; and cut into slices. The slices were baked, dewaxed, hydrated, immersed in hematoxylin solution for 3 min, differentiated in alcoholic hydrochloric acid for 15 s, washed, blued in bluing buffer for 15 s, washed, immersed in eosin solution for 3 min, washed, dehydrated, transparentized, mounted, and examined under a microscope (CX41, Olympus, Tokyo, Japan).
Micro-computed tomography (CT) scanning
The knee joints of the mice were scanned by using an X-ray microtomograph (micro-CT, SkyScan1174, Bruker, Kontich, Belgium) at 50 kV voltage, 800 µA current, 12 µm scanning resolution, and 1,304×1,024 field of view. Trabecular bone in the subchondral region of the tibia was set as the 3-dimensional (3D) reconstruction region of interest (ROI). The 3D image reconstruction was performed with N-Recon software (Bruker, Belgium), and 3D analysis was conducted with CT-AN software (Bruker, Belgium).
Reverse transcription polymerase chain reaction (RT-PCR)
We extracted RNA from the cells or articular cartilages and reverse transcribed into cDNA in accordance with the instructions of regents or kits. Fluorogenic quantitative PCR (CFX Connect, Bio-Rad, Hercules, CA, USA) was performed to amplify cDNA. The PCR reaction system (20 µL) consisted of RNase-free distilled water (7 µL), cDNA/DNA (1 µL), forward primer (1 µL), reverse primer (1 µL), and 2× UltraSYBR Mixture (10 µL). Table 1 lists the sequences of forward and reverse primers that were synthetized by General Biosystems (Anhui, China). The PCR parameters consisted of pre-denaturation for 10 min at 95 ℃, denaturation for 10 s at 95 ℃, annealing for 30 s at 58 ℃, elongation for 30 s at 72 ℃, and 40 cycles. Parameters for dissociation curve analysis consisted of 15 s at 95 ℃, 1 min at 58 ℃, 15 s at 95 ℃, 15 s at 8 ℃, and 15 s at 58 ℃. They were measured stepwise from 95 ℃ every 0.5 ℃. The products of PCR were evaluated on an RT-PCR system (CFX Connect, Bio-Rad, USA). We used GAPDH and U6 as internal controls. The relative expression of genes was calculated via the 2−ΔΔCt method (28).
Table 1
| Primers | Sequences | Length of primers (bp) | Length of products (bp) |
|---|---|---|---|
| lncRNA MEG3 | For: ATTATGAGGCAAATGGAGGC | 20 | 228 |
| Rev: TGGGTCAGGACAGGGAGTT | 19 | ||
| miR-34a | For: TGGCAGTGTCTTAGCTGGTTGT | ||
| Rev: universal primera | |||
| GAPDH | For: TCAACGGCACAGTCAAGG | 18 | 357 |
| Rev: TGAGCCCTTCCACGATG | 17 | ||
| U6 | For: GCTTCGGCAGCACATATACTAAAAT | 25 | 91 |
| Rev: CGCTTCACGAATTTGCGTGTCAT | 23 |
a, representative downstream primers are the primers that come with the kit. RT-PCR, reverse transcription polymerase chain reaction.
Western blot analysis
Western blot analysis was performed in accordance with a previous report (29). Cells or articular cartilages were lysed in lysis buffer at 4 ℃ for 30 min and centrifuged at 9,391 ×g for 10 min. The supernatant was carefully collected to obtain total protein. Protein concentration was examined with a bicinchoninic acid (BCA) kit. The protein was denatured, loaded to conduct sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) for 1–2 h, and transferred to a membrane for 30–50 min through a wet method. The membrane was then incubated in primary antibody buffer at 4 ℃ overnight, washed, and incubated in secondary antibody buffer at room temperature for 1–2 h. Enhanced chemiluminescent solution was added, and the membrane was evaluated on an imaging system (ChemiDoc XRS+, Bio-Rad, USA). The gray values of the blots were analyzed with Quantity One software (Bio-Rad, USA).
ELISA assay
In accordance with a previous report (30), the cells were seeded in 12-well plates at a density of 8×104 cells per well. After the cells were cultured until fully adherent, the culture media were replaced with complete media containing different concentrations of LPS (1, 3, 5, 7, and 9 µg/mL). After 24 or 48 h of culture, the cell supernatants were collected. The levels of IL-1β were determined with an IL-1β ELISA kit in accordance with the manufacturer’s instructions. Briefly, 40 µL of diluents and 10 µL of samples were mixed in a 96-well plate. Enzyme-conjugated reagents (100 µL) were then added to each well. The plate was sealed, incubated at 37 ℃ for 60 min, and washed 5 times with washing solution. After the addition of chromogenic reagents (50 µL of A and 50 µL of B) to each well, the plate was kept at 37 ℃ in the dark for 15 min. Stop buffer (50 µL) was added into each well, and the absorbance of each well was measured within 15 min at 450 nm on a microplate reader (Safire 2, Tecan, Switzerland). Then, the concentration of IL-1β was calculated.
Cell transfection
The lncRNA MEG3-pmir-GLO plasmid was constructed by General Biosystems (Anhui, China). When cell confluence reached approximately 70%, the culture media was replaced with 0.5 mL of serum-free media. Lipofectamine 3000 (2.5 µL) was diluted in 65.2 µL of Opti-MEM. Empty plasmid or the lncRNA MEG3-pmir-GLO plasmid (1.25 µg) and 2.5 µL of P3000 were mixed with 62.5 µL of Opti-MEM at room temperature for 5 min. Lipofectamine 3000 and the plasmid were then mixed at room temperature for 15 min. The mixture was added into the corresponding wells of 6-well plates which were then cultured at 37 ℃ and 5% CO2 for 6 h. Culture media containing 20% FBS (0.5 mL) were added, and the cells were cultured at 37 ℃ and 5% CO2 for another 48 h.
Cell grouping
The cells were divided into 4 groups: control, LPS, Vector + LPS, and lncRNA MEG3 + LPS. When the cells were fully adherent, complete media containing 7 µg/mL LPS were added. After culturing for 48 h, the cells were transfected with the empty plasmid (Vector group) or the lncRNA MEG3-pmir-GLO plasmid (lncRNA MEG3 group) as mentioned above. The levels of lncRNA MEG3 and miR-34a were examined through RT-PCR. The levels of Klotho, FGF23, Bcl-2, Bax, TGF-β1, Caspase 3, and Caspase 8 were examined by western blot analysis.
Statistical analysis
Data were expressed as mean ± standard deviation and analyzed with one-way analysis of variance (ANOVA) via SPSS software 20.0 (IBM Corp., Armonk, NY, USA). Differences were statistically significant when the P value was less than 0.05.
Results
Pathological and structural characterizations of the OA model
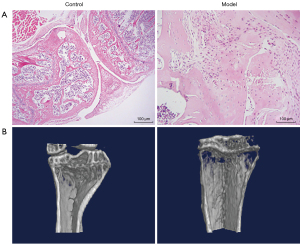
The pathological and structural characterizations of OA model mice were conducted through hematoxylin and eosin (HE) staining and micro-CT scanning, respectively. The resulting images are presented in Figure 1. As shown in Figure 1A, in the control mice, the cartilage surface of the femoral condyle and tibial plateau was smooth and intact. No cartilage was worn or broken, and the cartilage was thick. In the model mice, the medial cartilage surface of the tibial plateau was worn, but the articular surface remained intact, and no cartilage surface was broken. Local chondrocytes were reduced and hypertrophied, and cell arrangement was disordered. As depicted in Figure 1B, in the control mice, bone structures were normal, and bone surfaces were smooth. However, osteoporosis was found in the model mice.
Levels of miR-34a and Klotho in the OA model
The levels of miR-34a and Klotho in the OA model were examined by applying RT-PCR and western blot analyses and are shown in Figure 2. Compared with those in the control group, Klotho expression was significantly decreased and miR-34a expression was significantly increased (P<0.05) in the model group.

Screening of LPS concentration and treatment time
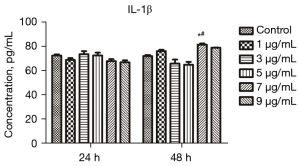
The IL-1β level in the supernatant of cells treated with different concentrations of LPS for different durations was measured through ELISA to screen the appropriate LPS concentration and treatment time. The results are provided in Figure 3. The IL-1β level after treatment with 7 µg/mL LPS for 48 h (P<0.05) in the treated mice was significantly elevated compared with that in the control group. The IL-1β level in the cells treated with 7 µg/mL LPS for 24 h was also significantly elevated (P<0.05) compared with that in the control group. Therefore, 7 µg/mL and 48 h were selected as the optimal concentration and treatment time for LPS, respectively, for the following experiments.
Transfection efficiency of the lncRNA MEG3 overexpression vector
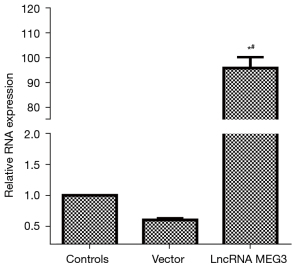
The RNA level of lncRNA MEG3 was detected via RT-PCR to verify the transfection efficiency of the lncRNA MEG3 overexpression vector. The results are shown in Figure 4. Compared with that in the control group, the expression of lncRNA MEG3 in the lncRNA MEG3 overexpression group was significantly up-regulated (P<0.05). The level of lncRNA MEG3 in the control group was similar to that in the Vector group. These results indicated that the lncRNA MEG3 overexpression vector was effective and transfected successfully.
Levels of lncRNA MEG3 and miR-34a
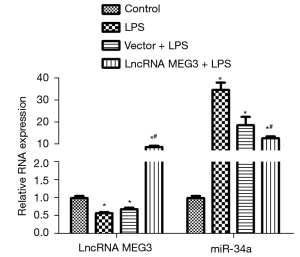
The levels of lncRNA MEG3 and miR-34a in the C28/I2 cells of various groups examined by RT-PCR are shown in Figure 5. Compared with those in the control group, lncRNA MEG3 expression decreased significantly and miR-34a expression increased sharply (P<0.05) in the LPS group. No significant difference in the levels of lncRNA MEG3 and miR-34a between the LPS group and the Vector + LPS group was observed. Interestingly, compared with those in the LPS group, lncRNA MEG3 expression increased remarkably and miR-34a expression decreased significantly (P<0.05) in the lncRNA MEG3 + LPS group.
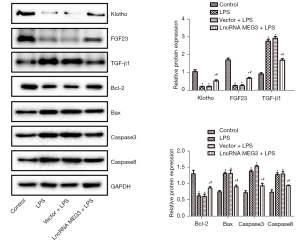
Levels of Klotho, FGF23, Bcl-2, Bax, TGF-β1, Caspase 3, and Caspase 8
The levels of Klotho, FGF23, Bcl-2, Bax, TGF-β1, Caspase 3, and Caspase 8 in the C28/I2 cells of various groups were determined through western blot analysis and are shown in Figure 6. Compared with those in the control group, the expression levels of Klotho, FGF23, and Bcl-2 in the LPS group were sharply decreased and the expression levels of Bax, TGF-β1, Caspase 3, and Caspase 8 were remarkably increased (P<0.05) in the treatment groups. The reverse transcription quantitative polymerase chain reaction (RT-qPCR) results provided in Figure 5 showed that the expression level of miR-34a in cells was very high. Although the vector itself might have an effect on miRNA, causing the inhibition of miR-34a expression, it did not affect the translation process. Therefore, in this protein detection experiment, no significant difference between the protein expression levels of Klotho, FGF23, Bcl-2, Bax, TGF-β1, Caspase 3, and Caspase 8 in the LPS group and those in the Vector + LPS group was observed. Interestingly, compared with those in the LPS group, the expression of Klotho, FGF23, and Bcl-2 was significantly elevated and the expression of Bax, TGF-β1, Caspase 3, and Caspase 8 were significantly reduced (P<0.05) in the lncRNA MEG3 + LPS group.
Discussion
In this study, C57BL/6 mice were used to establish an OA model. Pathological results showed that although the medial cartilage surface of the tibial plateau of the model mice had been worn, the articular surface remained intact, and no cartilage surface was broken. Moreover, decreased and hypertrophied local chondrocytes and disordered cell arrangement were found in the model mice. These were taken as pathological manifestations of OA. Moreover, micro-CT scanning found that the model mice exhibited osteoporosis. Therefore, the OA model in this study had been established successfully.
The glycolipid LPS is the main mediator of bacterial infection and disease evolution and plays a key role in inducing strong inflammatory responses, including the production of proinflammatory cytokines and chemokines (31). It is considered a key proinflammatory factor associated with OA (32). Previous studies have shown that LPS can induce chondrocyte inflammation damage in vitro (33) and promote the release of IL-1β (34). Another previous study revealed that LPS can notably induce chondrocyte damage, enhance cell apoptosis, and stimulate IL-1β release (35). In this study, ELISA was utilized to detect the level of IL-1β in the cell supernatant to screen the LPS concentration and treatment time suitable for C28/I2 chondrocytes. The level of IL-1β increased significantly when the LPS concentration was 7 µg/mL and the treatment time was 48 h. Therefore, we chose 48 h of treatment with 7 µg/mL LPS to establish a cell model. The cells were treated with LPS to induce the cellular inflammation reaction, and the mouse model was induced through papain injection. The inflammation reaction in these 2 cases shared a commonality. Papain is related to the IgE response (36), and it can promote the activation of the nuclear factor κB (NF-κB) pathway and nod-like receptor protein 3 (NLRP3) inflammasome (26). Changes in IgE levels can also be observed in LPS-induced inflammation (37), which is also related to NF-κB pathway activation and the NLRP3 inflammasome (38). Moreover, in this study, we found that the reactions in these 2 cases were consistent. Therefore, in our opinion, the cellular studies were closely connected with the mouse model.
In this study, we found that the expression of Klotho was significantly decreased and the expression of miR-34a was significantly increased in the OA model, suggesting that the formation of OA might be related to the changes in the expression of Klotho and miR-34a. In addition, we constructed the lncRNA MEG3 overexpression vector, which was successfully transfected into C28/I2 cells. After C28/I2 cells were treated with LPS, the expression levels of lncRNA MEG3 and Klotho decreased significantly, and the expression of miR-34a increased significantly. These results indicated that LPS damage to cells might be related to changes in the expression of lncRNA MEG3, Klotho, and miR-34a. Furthermore, after treatment with the lncRNA MEG3 overexpression vector, miR-34a expression was reduced and Klotho expression was enhanced, demonstrating that the up-regulation of lncRNA MEG3 could inhibit miR-34a expression and promote Klotho expression. Although further verification of the interaction between miR-34a and lncRNA MEG3 through dual luciferase experiments is needed to increase the validity of this study’s conclusion, the interaction between miR-34a and lncRNA MEG3 has been also reported previously (15,16). The results of this study were consistent with those of previous reports (15,16), indicating that lncRNA MEG3 could affect the expression of miR-34a.
The hormone FGF23 is an important regulator of serum phosphorus concentration (39). Serum phosphate is a key element in mineralization, and elevated serum FGF23 concentration is associated with mineralization defects, such as rickets (40). Although the serum calcium level is normal, the systemic FGF23 deletion caused by the mutation and inactivation of the FGF23 gene leads to the elevation of serum phosphorus levels and the calcification of tissue (40). When FGF23 is produced by osteocytes, it circulates in the serum, reaches the kidneys, and inhibits phosphate reabsorption (41). It also regulates other important cytokines that may be involved in mineralization through the direct action of Klotho/FGFR1 or other possible signal receptors in the cartilage area (42). The peptide factor TGF-β promotes cell growth and proliferation and regulates vascular endothelial cell growth, inflammatory cell chemotaxis, fibroblast proliferation, and extracellular matrix synthesis and degradation (43). Moreover, it plays an important role in immune regulation and tissue repair (42). Pombo-Suarez et al. reported that the expression of TGF-β1, TGF-β2, and TGF-β3 in the chondrocytes of OA was up-regulated by varying degrees and was consistent with the percentage of TGF-β-positive chondrocytes (44). Synovial hyperplasia, fibrosis, and osteophyte formation at the joint edge are promoted by TGF-β1 during the progress of OA (45). Consistent with the above reports (42,44), this study revealed that after LPS treatment, FGF23 expression decreased, whereas TGF-β1 expression increased in C28/I2 cells. The overexpression of lncRNA MEG3 could up-regulate FGF23 expression and down-regulate TGF-β1 expression. These results indicated that lncRNA MEG3 was capable of protecting chondrocytes injured by LPS through promoting FGF23 expression and inhibiting TGF-β1 expression.
Cell apoptosis controls a series of signal cascades in cells that cause cell death without initiating an inflammatory response. The proportion of Bcl-2 family members is a key factor in the regulation of cell apoptosis. While Bcl-2 has an antiapoptotic effect, Bax has a proapoptotic effect. In particular, the Bcl-2/Bax ratio is a molecular switch that initiates cell apoptosis and plays a key role in determining susceptibility to apoptosis (46). Caspase is an important effector molecule in cell apoptosis. Caspase 8 participates in the execution of apoptosis, induces the activation of Caspase 3, initiates a cascade reaction, and then causes apoptosis (47). The inhibition of Caspase 3 activation can effectively suppress the occurrence of apoptosis (48). This study found that after C28/I2 cells were treated with LPS, the expression of the proapoptotic proteins Bax, Caspase 3, and Caspase 8 increased, and the expression of the antiapoptotic protein Bcl-2 decreased. These results suggested that LPS would damage chondrocytes by up-regulating the expression of Bax, Caspase 3, and Caspase 8 by down-regulating the expression of Bcl-2. The overexpression of lncRNA MEG3 could depress the expression of Bax, Caspase, and Caspase 8 and boost the expression of Bcl-2, indicating that lncRNA MEG3 is capable of protecting chondrocytes by regulating the expression of apoptosis-related proteins. Although the expression of apoptosis-related molecules in the cells was determined, apoptosis was not examined in the cells and mouse models. The absence of apoptosis detection was a limitation of this study. We will detect apoptosis in animal models and cell models in a future study.
This study detected the expression of miR-34a and Klotho in a mouse model. Our idea was to first discover the phenomenon in animal models, then verify this phenomenon in cell models, and conduct preliminary research on the molecular mechanism of the phenomenon in cell models. Of course, if indicators, such as FGF23, TGF-β1, Bcl-2, Bax, Caspase 3, and Caspase 8, were also examined in animal models, the evidence would indeed show increased adequacy. Therefore, we intend to investigate the effects on the miR-34a/Klotho axis in animal models and cell models in detail in our next study.
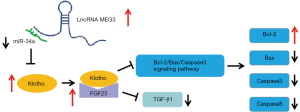
In conclusion, lncRNA MEG3 regulates the expression of FGF23, Bcl-2, Bax, TGF-β1, Caspase 3, and Caspase 8 by regulating the miR-34a/Klotho axis, thereby affecting the progress of OA. The signal flow that is regulated by lncRNA MEG3 in the progress of OA is illustrated in Figure 7. The lncRNA MEG3 and miR-34a might be diagnostic or prognostic markers and potential targets for OA treatment. Some reagents and drugs for the diagnosis and treatment of OA could be developed on the basis of lncRNA MEG3 and miR-34a.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-894/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-894/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-894/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Animal experiments were granted by the Ethics Committee of the First Affiliated Hospital of Jinan University (No. 20190550x), in compliance with the institutional guidelines of the First Affiliated Hospital of Jinan University for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fang H, Huang L, Welch I, et al. Early Changes of Articular Cartilage and Subchondral Bone in The DMM Mouse Model of Osteoarthritis. Sci Rep 2018;8:2855. [Crossref] [PubMed]
- Jahanban-Esfahlan R, Mehrzadi S, Reiter RJ, et al. Melatonin in regulation of inflammatory pathways in rheumatoid arthritis and osteoarthritis: involvement of circadian clock genes. Br J Pharmacol 2018;175:3230-8. [Crossref] [PubMed]
- Crossley KM, Schache AG, Ozturk H, et al. Pelvic and Hip Kinematics During Walking in People With Patellofemoral Joint Osteoarthritis Compared to Healthy Age-Matched Controls. Arthritis Care Res (Hoboken) 2018;70:309-14. [Crossref] [PubMed]
- Khan NM, Ahmad I, Haqqi TM. Nrf2/ARE pathway attenuates oxidative and apoptotic response in human osteoarthritis chondrocytes by activating ERK1/2/ELK1-P70S6K-P90RSK signaling axis. Free Radic Biol Med 2018;116:159-71. [Crossref] [PubMed]
- Rapicavoli NA, Qu K, Zhang J, et al. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife 2013;2:e00762. [Crossref] [PubMed]
- Kawasaki N, Miwa T, Hokari S, et al. Long noncoding RNA NORAD regulates transforming growth factor-β signaling and epithelial-to-mesenchymal transition-like phenotype. Cancer Sci 2018;109:2211-20. [Crossref] [PubMed]
- Zhang J, Li Z, Liu L, et al. Long noncoding RNA TSLNC8 is a tumor suppressor that inactivates the interleukin-6/STAT3 signaling pathway. Hepatology 2018;67:171-87. [Crossref] [PubMed]
- Li S, Yang J, Xia Y, et al. Long Noncoding RNA NEAT1 Promotes Proliferation and Invasion via Targeting miR-181a-5p in Non-Small Cell Lung Cancer. Oncol Res 2018;26:289-96. [Crossref] [PubMed]
- Xiong WC, Han N, Wu N, et al. Interplay between long noncoding RNA ZEB1-AS1 and miR-101/ZEB1 axis regulates proliferation and migration of colorectal cancer cells. Am J Transl Res 2018;10:605-17. [PubMed]
- Ren H, Yang X, Yang Y, et al. Upregulation of LncRNA BCYRN1 promotes tumor progression and enhances EpCAM expression in gastric carcinoma. Oncotarget 2018;9:4851-61. [Crossref] [PubMed]
- Jalali S, Bhartiya D, Lalwani MK, et al. Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS One 2013;8:e53823. [Crossref] [PubMed]
- Zhou X, Yuan P, Liu Q, et al. LncRNA MEG3 Regulates Imatinib Resistance in Chronic Myeloid Leukemia via Suppressing MicroRNA-21. Biomol Ther (Seoul) 2017;25:490-6. [Crossref] [PubMed]
- Tong P, Peng QH, Gu LM, et al. LncRNA-MEG3 alleviates high glucose induced inflammation and apoptosis of retina epithelial cells via regulating miR-34a/SIRT1 axis. Exp Mol Pathol 2019;107:102-9. [Crossref] [PubMed]
- Huang X, Gao Y, Qin J, et al. The mechanism of long non-coding RNA MEG3 for hepatic ischemia-reperfusion: Mediated by miR-34a/Nrf2 signaling pathway. J Cell Biochem 2018;119:1163-72. [Crossref] [PubMed]
- Lichner Z, Fendler A, Saleh C, et al. MicroRNA signature helps distinguish early from late biochemical failure in prostate cancer. Clin Chem 2013;59:1595-603. [Crossref] [PubMed]
- Zhao N, Lin T, Zhao C, et al. MicroRNA-588 is upregulated in human prostate cancer with prognostic and functional implications. J Cell Biochem 2017; Epub ahead of print. [Crossref] [PubMed]
- Bonetti P, Climent M, Panebianco F, et al. Dual role for miR-34a in the control of early progenitor proliferation and commitment in the mammary gland and in breast cancer. Oncogene 2019;38:360-74. [Crossref] [PubMed]
- Zhang L, Liao Y, Tang L. MicroRNA-34 family: a potential tumor suppressor and therapeutic candidate in cancer. J Exp Clin Cancer Res 2019;38:53. [Crossref] [PubMed]
- Endisha H, Datta P, Sharma A, et al. MicroRNA-34a-5p Promotes Joint Destruction During Osteoarthritis. Arthritis Rheumatol 2021;73:426-39. [Crossref] [PubMed]
- Chen H, Wang J, Hu B, et al. MiR-34a promotes Fas-mediated cartilage endplate chondrocyte apoptosis by targeting Bcl-2. Mol Cell Biochem 2015;406:21-30. [Crossref] [PubMed]
- Long FY, Shi MQ, Zhou HJ, et al. Klotho upregulation contributes to the neuroprotection of ligustilide against cerebral ischemic injury in mice. Eur J Pharmacol 2018;820:198-205. [Crossref] [PubMed]
- Nakahara T, Kawai-Koawase K, Matsui H, et al. Fibroblast growth factor 23 plays a protective role against vascular calcification. Eur Heart J 2013;34:4164. [Crossref]
- Chen K, Sun Z. Activation of DNA demethylases attenuates aging-associated arterial stiffening and hypertension. Aging Cell 2018;17:e12762. [Crossref] [PubMed]
- Fu T, Kemper JK. MicroRNA-34a and Impaired FGF19/21 Signaling in Obesity. Vitam Horm 2016;101:175-96. [Crossref] [PubMed]
- Liu Q, Ye J, Li S. Klotho Alleviates Renal Interstitial Fibrosis Via Inhibition of Endoplasmic Reticulum Stress in UUO Rats. Chinese Journal of Integrated Traditional and Western Nephrology 2015;16:203-6.
- Cheng F, Yan FF, Liu YP, et al. Dexmedetomidine inhibits the NF-κB pathway and NLRP3 inflammasome to attenuate papain-induced osteoarthritis in rats. Pharm Biol 2019;57:649-59. [Crossref] [PubMed]
- Zhai KF, Duan H, Chen Y, et al. Apoptosis effects of imperatorin on synoviocytes in rheumatoid arthritis through mitochondrial/caspase-mediated pathways. Food Funct 2018;9:2070-9. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Zhai KF, Zheng JR, Tang YM, et al. The saponin D39 blocks dissociation of non-muscular myosin heavy chain IIA from TNF receptor 2, suppressing tissue factor expression and venous thrombosis. Br J Pharmacol 2017;174:2818-31. [Crossref] [PubMed]
- Zhai KF, Duan H, Khan GJ, et al. Salicin from Alangium chinense Ameliorates Rheumatoid Arthritis by Modulating the Nrf2-HO-1-ROS Pathways. J Agric Food Chem 2018;66:6073-82. [Crossref] [PubMed]
- Tao H, Li N, Zhang Z, et al. Erlotinib Protects LPS-Induced Acute Lung Injury in Mice by Inhibiting EGFR/TLR4 Signaling Pathway. Shock 2019;51:131-8. [Crossref] [PubMed]
- Liu H, Pathak P, Boehme S, et al. Cholesterol 7α-hydroxylase protects the liver from inflammation and fibrosis by maintaining cholesterol homeostasis. J Lipid Res 2016;57:1831-44. [Crossref] [PubMed]
- Li W, Liu G, Wu X. PVT1 depletion protects cartilage ATDC5 cells against LPS-induced inflammatory injury by regulating the miR-24/ADAMTS5 axis. RSC Advances 2018;8:37518-27. [Crossref]
- Xiong XY, Liang J, Xu YQ, et al. The Tilapia collagen peptide mixture TY001 protects against LPS-induced inflammation, disruption of glucose metabolism, and aberrant expression of circadian clock genes in mice. Chronobiol Int 2019;36:1013-23. [Crossref] [PubMed]
- Konda VR, Desai A, Darland G, et al. META060 inhibits osteoclastogenesis and matrix metalloproteinases in vitro and reduces bone and cartilage degradation in a mouse model of rheumatoid arthritis. Arthritis Rheum 2010;62:1683-92. [Crossref] [PubMed]
- Iida H, Takai T, Hirasawa Y, et al. Epicutaneous administration of papain induces IgE and IgG responses in a cysteine protease activity-dependent manner. Allergol Int 2014;63:219-26. [Crossref] [PubMed]
- Hong SH, Ku JM, Kim HI, et al. Topical Application of KAJD Attenuates 2,4-Dinitrochlorobenzene-Induced Atopic Dermatitis Symptoms Through Regulation of IgE and MAPK Pathways in BALB/C Mice and Several Immune Cell Types. Front Pharmacol 2019;10:1097. [Crossref] [PubMed]
- Yu X, Lan P, Hou X, et al. HBV inhibits LPS-induced NLRP3 inflammasome activation and IL-1β production via suppressing the NF-κB pathway and ROS production. J Hepatol 2017;66:693-702. [Crossref] [PubMed]
- Agoro R, Montagna A, Goetz R, et al. Inhibition of fibroblast growth factor 23 (FGF23) signaling rescues renal anemia. FASEB J 2018;32:3752-64. [Crossref] [PubMed]
- Chen G, Liu Y, Goetz R, et al. α-Klotho is a non-enzymatic molecular scaffold for FGF23 hormone signalling. Nature 2018;553:461-6. [Crossref] [PubMed]
- Kamelian T, Saki F, Jeddi M, et al. Effect of Cholecalciferol therapy on serum FGF23 in vitamin D deficient patients: a randomized clinical trial. J Endocrinol Invest 2018;41:299-306. [Crossref] [PubMed]
- Donate-Correa J, Mora-Fernández C, Martínez-Sanz R, et al. Expression of FGF23/KLOTHO system in human vascular tissue. Int J Cardiol 2013;165:179-83. [Crossref] [PubMed]
- Meng X, Vander Ark A, Daft P, et al. Loss of TGF-β signaling in osteoblasts increases basic-FGF and promotes prostate cancer bone metastasis. Cancer Lett 2018;418:109-18. [Crossref] [PubMed]
- Pombo-Suarez M, Castaño-Oreja MT, Calaza M, et al. Differential upregulation of the three transforming growth factor beta isoforms in human osteoarthritic cartilage. Ann Rheum Dis 2009;68:568-71. [Crossref] [PubMed]
- Li Y, Golshirazian I, Asbury BJ, et al. Induction of high temperature requirement A1, a serine protease, by TGF-beta1 in knee joints of mouse models of OA. Histol Histopathol 2012;20:26-9. [Crossref]
- Zhang Z, Liang Z, Li H, et al. Perfluorocarbon reduces cell damage from blast injury by inhibiting signal paths of NF-κB, MAPK and Bcl-2/Bax signaling pathway in A549 cells. PLoS One 2017;12:e0173884. [Crossref] [PubMed]
- Pan YL, Han ZY, He SF, et al. miR 133b 5p contributes to hypoxic preconditioning mediated cardioprotection by inhibiting the activation of caspase 8 and caspase-3 in cardiomyocytes. Mol Med Rep 2018;17:7097-104. [Crossref] [PubMed]
- Zhao T, Fu Y, Sun H, et al. Ligustrazine suppresses neuron apoptosis via the Bax/Bcl-2 and caspase-3 pathway in PC12 cells and in rats with vascular dementia. IUBMB Life 2018;70:60-70. [Crossref] [PubMed]
(English Language Editor: J. Jones)


