
Screening and identification of osteoarthritis related differential genes and construction of a risk prognosis model based on bioinformatics analysis
Introduction
The incidence of osteoarthritis (OA) increases with age and has a prevalence in those over 65 years old of up to 50% (1). In recent years, OA pathogenesis is not a set of targets or that some gene interaction relationship has significantly changed, but a set of targets or some genes leading to related functional modules or biological network failure, including the enzymatic degradation of cartilage extracellular matrix, a lack of new extracellular matrix synthesis, cell death and apoptosis, abnormal activation of chondrocytes, and differentiation. Therefore, finding new and more meaningful biomarkers and the signaling pathways they participate in will play an important role in preventing and treating the occurrence and development of OA. With the rapid development of high-throughput technology, biological data has grown exponentially, and high-throughput biodetection technology represented by gene chips is widely used (2). This technology can obtain a large amount of disease-related gene information in a short time, making it possible to conduct a comprehensive and detailed analysis of diseases at the genetic level (3-5).
OA is a chronic inflammatory and degenerative joint disease characterized by cartilage degeneration, synovitis, and osteophyte formation, often leading to chronic disability in the elderly (6). While the current pathogenic mechanism of the disease is still unclear, most past research topics focused on cartilage tissue. With the continuous deepening of research, the role of synovial lesions in the occurrence and development of OA has attracted great attention. More than 90% of OA patients were found to have confirmed synovial lesions, and their degree was associated with severe pain and joint dysfunction (7). In addition, the occurrence of synovitis has also been reported, which may promote cartilage degeneration. While the search for the generating mechanisms of synovial lesions helps find precise therapeutic targets, fundamentally solving the problem of OA depends on the identification of biological markers (8-10). Therefore, finding disease differential genes and related signaling pathways is an urgent issue in the pathogenesis and therapeutic target research of the disease. With the rapid development of gene chip and RNA sequencing technologies, bioinformatics analysis has become an important research direction, providing new clues and core data for identifying reliable differentially expressed genes (DEGs), microRNAs (miRNAs), circular RNAs (cirRNAs), and long-chain non-coding RNAs (lncRNAs), and showing great advantages in screening candidate biomarkers for various diseases (11). However, since the samples originate from different sequencing platforms, the expressed messenger RNA (mRNA) results are inconsistent with the gene spectrum, and a large part of the bioinformatics analysis of OA is limited to a single chip data. While this may produce results which are limited and of poor reliability, multi-group microarray data can effectively solve this problem (12). Moreover, as many studies have progressed, large amounts of genetic information uploaded to public databases has not been effectively utilized (13). Related studies (14,15) only analyzed the expression and functional enrichment of OA differential genes, focusing more on the impact of OA risk analysis. However, few people have analyzed the differential genes in the prognosis of patients with OA and the construction of the risk prognosis model. In this study, the advantages of the previous two factors were skillfully combined to form a prognosis model for joint analysis of morbidity and prognosis. In addition, the data in the database included in this study are the latest research data, which has better effectiveness analysis.
Advantages of using bioinformatics techniques to analyze the incidence and prognosis of OA: (I) it is a combination of molecular biology and information technology to study biological problems by using the methods of applied mathematics, informatics, statistics, and computer science; (II) the data comes from the most authoritative database in the world, which is more credible and practical; (III) it is conducive to data sharing and academic exchange, etc.
Therefore, this study used multiple groups of previously used gene chip data to screen, analyze, and identify more biological meaningful markers and to provide new ideas in OA research. We present the following article in accordance with the TRIPOD reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1135/rc).
Methods
Data collection and preprocessing
Using “osteoarthritis” as the reference word, the corresponding gene chips were searched in the Gene Expression Omnibus (GEO) database of NCBI (https://www.ncbi.nlm.nih.gov/geo/), and four data sets meeting the experimental requirements were downloaded. The sequence numbers were GSE12021, GSE55235, GSE55457, and GSE55584, and all were from the GPL96 platform, with the chip type being an Affy-Metrix Human Genome U133a. The data set of the GSE12021 chip was submitted by Huber in 2010 (16) and included 13 normal synovial tissue samples, 20 OA tissue samples, and 20 synovial tissue samples of rheumatoid arthritis (RA). Data sets GSE55235, GSE55457, and GSE55584 were submitted by Woetzel et al. in 2014 (17), and included 20 normal synovial tissue samples, 26 OA synovial tissue samples, and 33 RA synovial tissue samples. In this study, normal synovial tissue samples and OA synovial tissue samples from four data sets were selected as samples for subsequent analysis. In this study, bioinformatics techniques were used to screen and identify differential genes in OA, functional enrichment analysis, protein-protein interaction (PPI) network analysis, prognostic critical genes analysis, and sensitivity and specificity analysis of area under the concentration-time curve (AUC).
Screening and analysis of DEGs
Data from GSE12021, GSE55235, GSE55457, and GSE55584 datasets were merged, and batch corrected using the “sva” analysis package in R, then analyzed using “limma” analysis package 8, where DEGs were selected with P<0.05, |log2fold change (FC)| >1. We investigated the functional roles of hub genes with a degree ≥10 and used the cBioPortal (https://www.cbioportal.org/) online platform to analyze gene networks and co-expressed genes. The biological network gene oncology tool (BiNGO) (version 3.0.4) in the Cytoscape plug-in was then used to visualize the hub gene biological process (BP).
Functional enrichment analysis of genes
Gene Ontology (GO) functional enrichment of differential gene expression and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. GO analysis of biological mechanisms was used for identification of high-throughput genomic or transcriptome data. This includes molecular function (MF), BP, and cellular component (CC). The KEGG database was used to identify functional and biological correlations of candidate targets. The “ClusterProfiler” enrichment package in R language was used to analyze the GO function and KEGG pathway enrichment of differential genes.
PPI network analysis
PPI network construction and key gene analysis imported all differential genes into the STRING online database (https://string-db.org/) for protein interaction analysis, and PPI was visualized using Cytoscape software. The Cytohubba plugin in the Cytoscape software was applied for the top 10 differential genes in the degree value (degree) as a key gene in the PPI. The DisGeNET database (https://www.disgenet.org/, version 5.0) is one of the platforms containing the genes and variants associated with human disease, and genes were retrieved in this database. The above targets were then successively imported into the database to obtain the target type information (protein class).
Analysis of key modules and functional enrichment analysis
PPI was analyzed by using Molecular COmplex DEtection (MCODE) in Cytoscape software. The standard setting: cut-off value of node score =0.2, K-core =2, degree cut-off =2, calculated the score value of MCODE. A score >6 was used as the screening criterion for significance modules, and the online database DAVID (https://david.ncifcrf.gov/) was used to conduct pathway enrichment analysis for genes in the selected modules.
Prognostic analysis of hub genes
The R software (version 4.0.2) is a collection of toolkits used for the annotation, processing, analysis, and visualization of biological data and consists of a series of packages. We retrieved our transcriptome data and the clinical group data of the screened hub genes in The Cancer Genome Atlas (TCGA) database. We then ran the analysis using R software to obtain the relevant mRNA expression of the mRNA of hub genes and mapped its corresponding risk curve, which represents the risk analysis of the prognosis of OA. In addition, we ran independent prognostic analysis and trend analysis in the R language environment. The independent prognostic analysis was expressed in the form of forest diagrams, while the trend analysis was expressed by the receiver operating characteristic (ROC) curves. Single-factor and multi-factor Cox regression analysis indicate the clinical staging, and the risk score represents an independent prognostic factor.
Statistical analysis
SPSS (23.0) statistical software was used for data analysis. The measurement data were expressed as mean ± standard deviation () and tested by one-way analysis of variance (ANOVA). P<0.05 indicated that the difference was statistically significant. In this study, variance analysis was performed on the relevant data of the OA test group and the normal healthy volunteers control group. In the univariate Cox regression, multivariate Cox regression and ROC analysis, t test was required for the selected data.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Screening and identification of differential genes
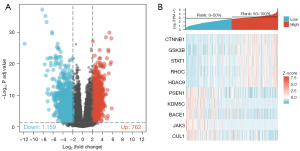
The dataset of GSE12021, GSE55235, GSE55457, and GSE55584 contained 33 normal synovial tissue samples and 40 OA synovial tissue samples. A total of 1,921 differential genes were screened, including 762 up-regulated genes and 1,159 down-regulated genes (Figure 1A), and heat maps visually show differences in gene expression between OA patients and healthy controls (Figure 1B).
PPI network construction and module analysis
We built the PPI network of DEGs using Cytoscape software and used its plug-in MCODE to obtain the most important modules. Results of the function analysis of the hub genes using DAVID showed the selected DEGs were mainly enriched in cell division, mitosis, nuclear division, and cell cycle, as shown in Table 1.
Table 1
| Term | Count | % | P value | FDR |
|---|---|---|---|---|
| hsa04610:Complement and coagulation cascades | 13 | 0.015564575 | 1. UE-05 | 0.014323205 |
| hsa04512:ECM-receptor interaction | 13 | 0.015564575 | 1.20E-04 | 0.154467703 |
| hsa04974:Protein digestion and absorption | 13 | 0.015564575 | 1.35E-04 | 0.172767737 |
| hsa04U5:p53 signaling pathway | 9 | 0.010775475 | 0.00415009 | 5.202475569 |
| hsa04510:Focal adhesion | 17 | 0.020353675 | 0.006182138 | 7.657719001 |
| hsa04110:Cell cycle | 12 | 0.0143673 | 0.008516724 | 10.40605319 |
| hsa05202:Transcriptional misregulation in cancer | 14 | 0.01676185 | 0.012777216 | 15.22807973 |
| hsa04151:PI3K-Akt signaling pathway | 23 | 0.027537325 | 0.014475026 | 17.08206542 |
| hsa05146:Amoebiasis | 10 | 0.01197275 | 0.021524457 | 24.38706471 |
| hsa04514:Cell adhesion molecules | 12 | 0.0143673 | 0.021828745 | 24.6885954 |
| hsa04114:Oocyte meiosis | 10 | 0.01197275 | 0.028056965 | 30.62192317 |
| hsa04068:FoxO signaling pathway | 11 | 0.013170025 | 0.035073485 | 36.78815863 |
| hsa04270:Vascular smooth muscle contraction | 10 | 0.01197275 | 0.037600738 | 38.88239916 |
| hsa05166:HTLV-I infection | 17 | 0.020353675 | 0.03777001 | 39.02035566 |
| hsa05144:Malaria | 6 | 0.00718365 | 0.040274546 | 41.02828533 |
| hsa03320:PPAR signaling pathway | 7 | 0.008380925 | 0.044757247 | 44.47065359 |
| hsa04914:Progesterone-mediated oocyte | 8 | 0.0095782 | 0.051422539 | 49.24770846 |
| hsa05200:Pathways in cancer | 23 | 0.027537325 | 0.05218087 | 49.76649163 |
| hsa04614:Renin-angiotensin system | 4 | 0.0047891 | 0.057734986 | 53.41962981 |
| hsa04611:Platelet activation | 10 | 0.01197275 | 0.065349769 | 58.03077189 |
| hsa04670:Leukocyte transendothelial migration | 9 | 0.010775475 | 0.077846058 | 64.69536318 |
| hsa04925:Aldosterone synthesis and secretion | 7 | 0.008380925 | 0.09381443 | 71.79207124 |
GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially expressed genes; FDR, false discovery rate.
GO and KEGG pathway enrichment analysis
The function and pathway enrichment analysis of hub genes was performed using DAVID. The results of GO analysis showed changes in the BP terms of the hub genes were significantly increased in signal pathways, regulation of cell division, regulation of complement activation, and mitotic cell cycle. On the other hand, changes in the CC terms were concentrated in the cytoplasmic area, extracellular area of the cell membrane, area around the nucleus of the cytoplasm, concentrated chromosomal centromeres, and platelets in the hyaluronic acid granules. GO enrichment analysis showed 151 differential genes, including dioxase activity, iron ion binding, cell adhesion, and ubiquitin protein ligase collection, and 216 items were enriched for downregulated differential genes, mainly including inhibition of transport factor binding, nucleotide sugar transmembrane transport activity, and hormone receptor binding (Figure 2).

PPI and key gene analysis
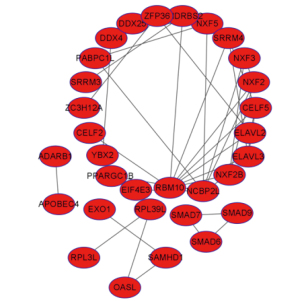
The STRING database (https://string-db.org) was used to construct PPI networks, and the threshold for interaction was set at 0.7. The filtered differential genes were imported into the STRING online database to obtain the PPI of the differential genes, and the network contained 313 nodes with 2,160 edges, 167 up-regulated genes, and 144 downregulated genes. The Cytohubba plug-in of Cytoscape software was applied to analyze the topological parameters of the network and indicate the importance of the gene in the network in the size of degree value. If the degree increased, the more genes were associated with it, and the more important the gene (Figure 3).
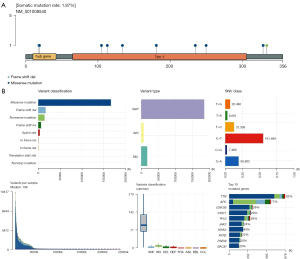
Landscape analysis of mutated genes in OA
Landscape analysis of mutant genes is a complex single-sample analysis, which can be used to study the mutation situation of a certain gene in the disease, including the physical location of mutation, panoramic waterfall map of mutation type, and the further analysis of mutation subgroups. We downloaded OA related gene mutation data, transcriptome data, and clinical data from TCGA database, and downloaded and visualized somatic mutations in patients using the MAf Tools software package in R software. A horizontal histogram showed patients with OA had a high frequency of gene mutation, and we found the mutation rate of OA susceptibility genes was 1.87%, suggesting patients with a high expression of OA susceptibility genes had a certain risk of disease. Missense mutations were the most significant in the 536 samples of patients with OA, in which the mutations of OA susceptibility genes were related to mononucleotide mutations, focusing on nonsense mutations, insertion mutations, and deletion mutations (Figure 4).
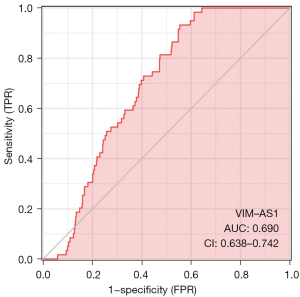
Evaluation of prognostic model of OA (ROC curve analysis)
The ROC results, with the abscissa as false positive rate (FPR) and the ordinate as true positive rate (TPR), were used to analyze the diagnostic efficacy of OA susceptibility genes in the pathogenesis of OA. The AUC (0.690) of the OA susceptibility gene, indicating that the HUB gene may be a potential diagnostic molecule. The area values under the ROC curve are between 0.5 and 1, and the closer the AUC is to 1, the better the diagnostic effect. The accuracy of the AUC ranged from 0.5 to 0.7, while that of the AUC ranged from 0.7 to 0.9. The accuracy of AUC was higher than 0.9, The risk prognosis model of patients with OA constructed in this study has high feasibility, good predictive ability, and certain clinical practicability (AUC =0.690) (Figure 5).
Discussion
OA is the most common total joint disease. While obesity, age, excessive exercise, inflammation, heredity. and trauma are closely related to its development, the etiology and pathogenesis of OA are unclear (18). Analysis of the underlying pathogenesis of OA is important for diagnosis and prognosis and the identification of drug therapy targets (19). As high-throughput sequencing and microarray technology can simultaneously provide information on the expression levels of thousands of genes in the human genome, this approach has been widely used to predict potential diagnostic and therapeutic targets for OA (20).
Differential gene analysis in this study shows the complexity of the pathogenesis of OA, which is not a simple single gene target acting alone, but a complex result of multiple gene target interactions (21). KEGG pathway enrichment results showed these differential genes were mainly involved in the MAPK signaling pathway, fatty acid metabolism pathway, apoptosis pathway, JAK-STAT signaling pathway, primary immune pathway, and hematopoietic cell lineage pathway. The MAPK signaling pathway can regulate chondrocyte proliferation, apoptosis, extracellular matrix metabolism, inflammatory factor secretion, and other processes, and play an important role in the pathological process of OA. Khan et al. (22) showed that the 3-phosphoinositol-dependent protein kinase (PDK) can promote chondrocyte apoptosis in OA through the p38 MAPK signaling pathway (23,24), while Yang et al. (25) found DUal-specificity phosphatase (DUSP) overexpression in OA inhibited the activation of MAPK signaling and the expression of the OA-associated matrix. Zhang et al. (26) revealed the JAK/STAT signaling pathway was regulated by significantly upregulated by proinflammatory cytokine gene expression, while MMP-13 expression in the IL-1-treated human chondrosarcoma cell lines induced JAK2 and STAT1/STAT2 activation and MMP-13 gene expression which was blocked by the pan-tyrosine kinase inhibitor AG490 (27-30). However, it was also found that treatment of this chondrocyte line with IL-1 also activated p38-MAPK.
GO analysis showed that down-regulated DEGs were mainly enriched in vascular development. Vascular endothelial growth factor A (VEGF A) is an important regulatory factor in bone development, and its expression was significantly increased in articular cartilage and synovium in advanced OA (31). VEGF A is involved in OA cartilage degeneration, osteophyte formation, sclerosis of subchondral bone cysts, and specific disease processes, such as, synovitis. Further, in cooperation with the IL-1β of VEGF A can significantly reduce the aggregation of proteoglycan and type II collagen gene expression and protein level, and inhibition of VEGF signaling can delay OA progression (32). Interestingly, recent studies (33-35) suggest the VEGF A signaling pathway has a dual function, which can promote angiogenesis and induce vascular degeneration. Our results showed that the VEGF A gene had low expression in the synovium of OA. We suspect this may be because the overexpression of VEGF protein in the body reversed the expression of the VEGF A gene, resulting in gene down-regulation. At the same time, it cannot be ruled out that the inconsistent results may be related to different gene pool selection. In conclusion, VEGF A plays an important role in the occurrence and development of OA, and its specific mechanism needs to be further explored (36).
PPI network analysis of these differential genes was carried out using the STRING online database, and the key target located in the center was obtained, in which, STAT1 is an indispensable component in regulating the inflammatory response of macrophages (37). If activated STAT1 is phosphorylated and translocated to the genes that regulate inflammation in the nucleus, JAK3, as a non-receptor tyrosine kinase, interacts with the common γ chain (IL2RG) of the cytokine receptor complex.
In conclusion, this study revealed CTNNB1 and GSK3B, as well as the STAT1, RHOC, HDAC9, PSEN1, KDM5C, BACE1, and JAK3 hub genes including CUL1, may play an important role in OA, and these biological markers may become drug targets and diagnostic markers of the disease. This study provides new insights into the molecular biology of OA and paves the way for the discovery of new biomarkers for its diagnosis.
Acknowledgments
Funding: This work was supported by the Zhongnan Hospital of Wuhan University Medical Science and Technology Innovation Platform Project (No. PTXM2020033).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1135/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1135/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Xiong Y, Mi BB, Liu MF, et al. Bioinformatics Analysis and Identification of Genes and Molecular Pathways Involved in Synovial Inflammation in Rheumatoid Arthritis. Med Sci Monit 2019;25:2246-56. [Crossref] [PubMed]
- Ren C, Li M, Du W, et al. Comprehensive Bioinformatics Analysis Reveals Hub Genes and Inflammation State of Rheumatoid Arthritis. Biomed Res Int 2020;2020:6943103. [Crossref] [PubMed]
- Xi Y, Fowdur M, Liu Y, et al. Differential expression and bioinformatics analysis of circRNA in osteosarcoma. Biosci Rep 2019;39:BSR20181514. [Crossref] [PubMed]
- Zou R, Zhang D, Lv L, et al. Bioinformatic gene analysis for potential biomarkers and therapeutic targets of atrial fibrillation-related stroke. J Transl Med 2019;17:45. [Crossref] [PubMed]
- Li F, Guo P, Dong K, et al. Identification of Key Biomarkers and Potential Molecular Mechanisms in Renal Cell Carcinoma by Bioinformatics Analysis. J Comput Biol 2019;26:1278-95. [Crossref] [PubMed]
- Huang PY, Wu JG, Gu J, et al. Bioinformatics analysis of miRNA and mRNA expression profiles to reveal the key miRNAs and genes in osteoarthritis. J Orthop Surg Res 2021;16:63. [Crossref] [PubMed]
- Ding J, Liang Z, Feng W, et al. Integrated Bioinformatics Analysis Reveals Potential Pathway Biomarkers and Their Interactions for Clubfoot. Med Sci Monit 2020;26:e925249. [Crossref] [PubMed]
- Li P, Che X, Gao Y, et al. Proteomics and Bioinformatics Analysis of Cartilage in Post-Traumatic Osteoarthritis in a Mini-Pig Model of Anterior Cruciate Ligament Repair. Med Sci Monit 2020;26:e920104. [Crossref] [PubMed]
- Wang S, Sun J, Yang H, et al. Profiling and bioinformatics analysis of differentially expressed circular RNAs in human intervertebral disc degeneration. Acta Biochim Biophys Sin (Shanghai) 2019;51:571-9. [Crossref] [PubMed]
- Wu F, Jiang X, Wang Q, et al. The impact of miR-9 in osteosarcoma: A study based on meta-analysis, TCGA data, and bioinformatics analysis. Medicine (Baltimore) 2020;99:e21902. [Crossref] [PubMed]
- Huang R, Meng T, Zhu R, et al. The Integrated Transcriptome Bioinformatics Analysis Identifies Key Genes and Cellular Components for Spinal Cord Injury-Related Neuropathic Pain. Front Bioeng Biotechnol 2020;8:101. [Crossref] [PubMed]
- Lin T, Chen W, Yang P, et al. Bioinformatics analysis and identification of genes and molecular pathways in steroid-induced osteonecrosis of the femoral head. J Orthop Surg Res 2021;16:327. [Crossref] [PubMed]
- Kou J, Liu G, Liu X, et al. Profiling and Bioinformatics Analysis of Differentially Expressed circRNAs in Spinal Ligament Tissues of Patients with Ankylosing Spondylitis. Biomed Res Int 2020;2020:7165893. [Crossref] [PubMed]
- Zhang Q, Zhang Y, Yan M, et al. Bioinformatics Analysis of the Molecular Mechanism of Late-Stage Heterotopic Ossification. Biomed Res Int 2020;2020:5097823. [Crossref] [PubMed]
- Wu J, Liu L, Hu H, et al. Bioinformatic analysis and experimental identification of blood biomarkers for chronic nonunion. J Orthop Surg Res 2020;15:208. [Crossref] [PubMed]
- Huber PW, Zhao WM. Detection of protein-RNA complexes in Xenopus oocytes. Methods 2010;51:82-6. [Crossref] [PubMed]
- Woetzel D, Huber R, Kupfer P, et al. Identification of rheumatoid arthritis and osteoarthritis patients by transcriptome-based rule set generation. Arthritis Res Ther 2014;16:R84. [Crossref] [PubMed]
- Lopes ECP, Paim LR, Matos-Souza JR, et al. Bioinformatics analysis of circulating miRNAs related to cancer following spinal cord injury. Biosci Rep 2019;39:BSR20190989. [Crossref] [PubMed]
- Jiang C, Li Z, Wu Z, et al. Integrated Bioinformatics Analysis of Hub Genes and Pathways Associated with a Compression Model of Spinal Cord Injury in Rats. Med Sci Monit 2020;26:e927107. [Crossref] [PubMed]
- Zhou J, Li C, Yu A, et al. Bioinformatics analysis of differentially expressed genes involved in human developmental chondrogenesis. Medicine (Baltimore) 2019;98:e16240. [Crossref] [PubMed]
- Huang K, Lin B, Liu Y, et al. Genomic Expression Profiling and Bioinformatics Analysis of Chronic Recurrent Multifocal Osteomyelitis. Biomed Res Int 2021;2021:6898093. [Crossref] [PubMed]
- Khan NM, Clifton KB, Lorenzo J, et al. Comparative transcriptomic analysis identifies distinct molecular signatures and regulatory networks of chondroclasts and osteoclasts. Arthritis Res Ther 2020;22:168. [Crossref] [PubMed]
- Yang B, Dong K, Guo P, et al. Identification of Key Biomarkers and Potential Molecular Mechanisms in Oral Squamous Cell Carcinoma by Bioinformatics Analysis. J Comput Biol 2020;27:40-54. [Crossref] [PubMed]
- Ma Y, Guo J, Li D, et al. Identification of potential key genes and functional role of CENPF in osteosarcoma using bioinformatics and experimental analysis. Exp Ther Med 2022;23:80. [Crossref] [PubMed]
- Yang F, Zhai Z, Luo X, et al. Bioinformatics identification of key candidate genes and pathways associated with systemic lupus erythematosus. Clin Rheumatol 2020;39:425-34. [Crossref] [PubMed]
- Zhang Z, Wang Q, Li Y, et al. Hub Genes and Key Pathways of Intervertebral Disc Degeneration: Bioinformatics Analysis and Validation. Biomed Res Int 2021;2021:5340449. [Crossref] [PubMed]
- Li F, Jin Y, Pei X, et al. Bioinformatics analysis and verification of gene targets for renal clear cell carcinoma. Comput Biol Chem 2021;92:107453. [Crossref] [PubMed]
- Ren Z, Li C, Gan Y, et al. Long noncoding RNA taurine-up regulated gene 1 for the prognosis of osteosarcoma: A protocol for meta-analysis and bioinformatics analysis. Medicine (Baltimore) 2021;100:e26182. [Crossref] [PubMed]
- Wang J, Gong M, Xiong Z, et al. Bioinformatics integrated analysis to investigate candidate biomarkers and associated metabolites in osteosarcoma. J Orthop Surg Res 2021;16:432. [Crossref] [PubMed]
- Sun F, Zhou JL, Peng PJ, et al. Identification of Disease-Specific Hub Biomarkers and Immune Infiltration in Osteoarthritis and Rheumatoid Arthritis Synovial Tissues by Bioinformatics Analysis. Dis Markers 2021;2021:9911184. [Crossref] [PubMed]
- Liu T, Huang J, Xu D, et al. Identifying a possible new target for diagnosis and treatment of postmenopausal osteoporosis through bioinformatics and clinical sample analysis. Ann Transl Med 2021;9:1154. [Crossref] [PubMed]
- Chang L, Yao H, Yao Z, et al. Comprehensive Analysis of Key Genes, Signaling Pathways and miRNAs in Human Knee Osteoarthritis: Based on Bioinformatics. Front Pharmacol 2021;12:730587. [Crossref] [PubMed]
- Wang JS, Duan MY, Zhong YS, et al. Investigating age-induced differentially expressed genes and potential molecular mechanisms in osteosarcoma based on integrated bioinformatics analysis. Mol Med Rep 2019;19:2729-39. [Crossref] [PubMed]
- Li Q, Liang J, Chen B. Identification of CDCA8, DSN1 and BIRC5 in Regulating Cell Cycle and Apoptosis in Osteosarcoma Using Bioinformatics and Cell Biology. Technol Cancer Res Treat 2020;19:1533033820965605. [Crossref] [PubMed]
- Carlson AK, Rawle RA, Wallace CW, et al. Global metabolomic profiling of human synovial fluid for rheumatoid arthritis biomarkers. Clin Exp Rheumatol 2019;37:393-9. [PubMed]
- Meng F, Du N, Xu D, et al. Bioinformatics Analysis of the Molecular Mechanism and Potential Treatment Target of Ankylosing Spondylitis. Comput Math Methods Med 2021;2021:7471291. [Crossref] [PubMed]
- Lee KI, Gamini R, Olmer M, et al. Mohawk is a transcription factor that promotes meniscus cell phenotype and tissue repair and reduces osteoarthritis severity. Sci Transl Med 2020;12:eaan7967. [Crossref] [PubMed]
(English Language Editor: B. Draper)


