
Hyperthermia enhances the efficacy of chemotherapeutic drugs in heat-sensitive cells through interfering with DNA damage repair
Introduction
At present, thermochemotherapy has been used in a small range in the clinical treatment of some cancers, including for the treatment of non-muscle invasive bladder cancer, rectal cancer, non-small cell lung cancer and ovarian cancer (1-4). Ovarian cancer is a leading cause of malignancy-related deaths in women worldwide. In Western countries, ovarian cancer is the 8th most common tumor (5). Clinically, most ovarian tumor tissues spread in the abdominal cavity. Therefore, hyperthermic intraperitoneal chemotherapy (HIPEC), as an aggressive form of regional chemotherapy, has gained much attention (6). Sukovas et al. demonstrated that hyperthermia (42–43 ℃) combined with cisplatin is more cytotoxic to the human ovarian adenocarcinoma cell line OVCAR-3 compared to single agent treatments (7). However, HIPEC is not effective for certain types of ovarian cancers. Tumor cells develop different mechanisms to protect themselves from extreme temperature, which is referred to as thermotolerance (8). Hatakeyama et al. identified hyperthermia-resistant cells, including SKOV3, ES2, HeyA8, PEO4, and KLE, and hyperthermia-sensitive cells, including A2780, A2780CP20, Hec-1A, SKUT-2, and ISHIKAWA. In the study, the median lethal temperature 50 (LT50) was used to distinguish cells, cells with LT50s above the median are heat-resistant cells, below the median, are heat-sensitive cells (9). However, the mechanisms by which hyperthermia enhances the effect of chemotherapeutic drugs is unclear.
Sukovas et al. reported that hyperthermia combined with cisplatin was more effective in suppressing glutamate dehydrogenase (GDH) activity and expression, and mitochondrial function, thereby limiting the survival of cancer cells (7) a previous study has also shown that heat enhances the uptake of drugs by cancer cells, leading to increased intracellular drug concentration. In addition, heat can increase the binding of drugs to DNA specific sites and thereby aggravate DNA damage and inhibit DNA repair, which can enhance the cytotoxic activity of drugs (10). However, the mechanisms underlying the therapeutic effects of thermochemotherapy remain to be fully elucidated. Luzhin et al. used mass spectrometry (MS)-based proteomics to demonstrate that hyperthermia can induce chromatin trapping of essential DNA damage repair factors, resulting in DNA damage repair dysfunction (11). However, this study did not involve the changes of related molecules in ovarian cancer cells, so our research mainly showed the changes of differential proteins in ovarian cancer cells after receiving thermochemotherapy This current study identified the mechanisms and explored the key molecules that enhance the sensitivity of heat-resistant cells to hyperthermia through quantitative proteomics.
Proteomics techniques, including liquid chromatography-mass spectrometry (LC-MS), have been widely used to explore the molecular protein markers and mechanisms associated with diseases, such as mental illnesses, cardiovascular diseases, and cancer, with good detection and reproducibility (12-16). In this investigation, heat-sensitive human ovarian cancer A2780 cell lines were treated with hyperthermia and chemotherapy. Cells were divided into 4 groups, with duplicates in each group. Samples were subjected to peptide extraction and the tandem mass tag (TMT)-labeled peptides were classified and analyzed with LC-MS. The differentially expressed proteins between groups were analyzed and verified to identify the key molecules involved in thermochemotherapy. These findings will promote the development of small molecule drugs through the discovery of the thermochemotherapy mechanism. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-955/rc).
Methods
Cell sample preparation
The human ovarian cancer cell line A2780 was purchased from Wuhan Punuosai Life Technology Co., Ltd. (Wuhan, China) and seeded in 100 mm Petri dishes until 60% confluency was reached. Cell was then treated as follows: control containing only culture media (group A); oxaliplatin (5 µmol/L) as the chemotherapeutic drugs (group B); heat stimulation at 42 ℃ for 1 hour (group C); or oxaliplatin in combination with heat stimulation (group D). Experiments were performed in duplicates. After 24 hours, the cells were collected and counted to ensure there were more than 1×107 cells per sample. Cells were then digested and placed in liquid nitrogen for rapid freezing for 10–20 seconds. Samples were stored at −80 ℃ until proteomics analysis.
Protein extraction
Samples were sonicated in lysis buffer (1% protease inhibitor, 8 mM urea) at 4 ℃, followed by centrifugation at 12,000 g for 10 minutes. The supernatant was collected and the concentration was measured.
Trypsin digestion
The protein supernatant was incubated with 5 mM DTT for 30 minutes at 56 ℃, followed by incubation with 11 mM iodoacetamide at room temperature in the dark for 15 minutes. The final concentration of urea was less than 2 M. Samples were then digested in pancreatin (1:50) at 37 ℃ overnight. A second digestion with pancreatin (1:100) was performed for 4 hours.
TMT/isobaric tagging for relative and absolute quantification (iTRAQ)
The peptides were desalted with Strata X C18 (Phenomenex) and freeze-dried in vacuo. A TMT kit/iTRAQ kit was used to tag the samples, which were reconstituted with 0.5 mM triethylammonium bicarbonate buffer (TEAB). The TMT/iTRAQ reagent was dissolved in acetonitrile and incubated with the peptide for 2 hours at room temperature. Finally, the tagged peptides were collected, the salt was removed, and the samples were dried by centrifugation in a vacuum environment.
High performance liquid chromatography (HPLC) fractionation
The peptides were separated with a gradient of 8% to 32% acetonitrile (pH 9.0) over 60 minutes into 60 fractions. The peptides were pooled into 14 components and vacuum freeze-dried for subsequent experiments.
LC-MS/MS analysis
Peptides were dissolved in 0.1% (v/v) aqueous formic acid solution (solvent A) and separated with extra high property liquid system (EASY-nLC 1000) at a stable flow rate of 350 nL/min. Subsequently, the liquid gradient of solvent B, which consists of 0.1% formic acid and 90% acetonitrile, was increased from 9% to 26% within the first 40 minutes, to 35% within the next 14 minutes, and to 80% in the next 3 minutes. The gradient was maintained at 80% for the last 3 minutes.
The separated peptides were injected into the nanospray ionization (NSI) source for ionization and analyzed using Q Exactive MS. The ion source voltage was set to 2.1 kV. The peptide precursor ions and their secondary fragments were detected and analyzed with high-resolution Orbitrap. The top 20 highest signal intensity peptide precursor ions to enter the HCD collision cell were selected and separated into 28% fragmentation energy separate parts after the first-level scan. The secondary MS analysis method is identical.
The quantitative value of each sample peptide was measured using MS. Since each protein corresponds to multiple peptides, the specific peptide corresponding to the protein was used to calculate the quantitative value. For each repeated experiment, the ratio of the quantitative protein value between two different groups was taken as the differential expression of the comparison group (ratio). The average of the two repeated ratio values was taken as the ratio of the compared groups. When the coefficient of variation (CV)-value <0.1 and the ratio >1.2 the peptides were considered to be upregulated. When <1/1.2, the peptides were considered to be downregulated.
Gene Ontology (GO) enrichment analysis
GO uses the UniProt-GOA database to classify proteins (http://www.ebi.ac.uk/GOA/). The UniProt ID was used to transform the protein ID to the GO ID, which was then used to retrieve the relevant information from the UniProt-GOA database. Where relevant protein information could not be searched using the UniProt-GOA database, InterProScan, a protein sequence-based algorithm software, was used to forecast the possible GO function of the protein.
Enrichment-based clustering
The functional information and the relevant enriched P values of the protein categories were gathered and the enriched cluster categories were screened (P<0.05). The filtered P value matrixes were transformed using the function x = –log10 (P value) and Z transformations were applied. Finally, the set of data obtained after Z transformation was analyzed with the hierarchical clustering method (Euclidean distance, average connection clustering) for unilateral clustering analysis. The relationship of the clustering was visually displayed using heat maps drawn using the function “heatmap.2” in the R language package “gplots”.
Western blot
Cell lysates were separated on 10% sodium dodecyl-sulfate (SDS)-polyacrylamide gels and transferred to polyvinylidene difluoride membranes. Membranes were probed using the primary antibodies against retinoblastoma-like 1 (RBL-1; p107 ab236518, 1:2,000) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Beyotime AG109 1:1,000), followed by incubation with secondary antibodies [horseradish peroxidase-labeled goat anti-mouse IgG (H+L); Beyotime; 1:1,000]. Protein bands were visualized using a multicolor fluorescence chemiluminescence imaging analysis system (Alpha America).
Statistical analysis
Two replicate experiments were performed in this study, Fisher’s exact test and t-test were used, and a P value <0.05 was considered statistically significant.
Results
Protein quantitative analysis
Hyperthermia mainly increased the sensitivity of the heat-sensitive ovarian cancer cells to chemotherapeutic drugs by inhibiting the repair of damaged DNA (9). In this study, ovarian cancer A2780 cells were treated with control media (group A), oxaliplatin alone (group B), hyperthermia at 42 ℃ (group C), or oxaliplatin and hyperthermia at 42 ℃ (group D). The peptides extracted from the cells were used for LC-MS analysis, followed by database search and biometric analysis. A 252,721 secondary spectra was obtained, including 102,016 usable effective spectra and the spectrum utilization rate was 40.4%. Through spectral analysis, a total of 54,841 peptides were identified, of which there were 53,166 specific peptides (Figure 1, Table 1).
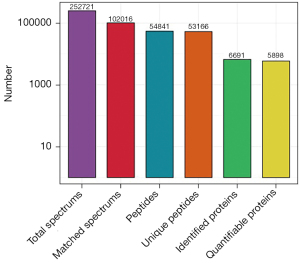
Table 1
| Total spectrum: 252,721 |
| Matched spectrum: 102,016 (40.4%) |
| Peptides: 54,841 |
| Unique peptides: 53,166 |
| Identified proteins: 6,691 |
| Quantifiable proteins: 5,898 |
MS, mass spectrometry.
Differential protein analysis between groups
The differential proteins expressed in the 4 treatment groups were analyzed. The quantitative value of each sample peptide was achieved using MS. Comparing hyperthermia-treated cells (group C) and control cells (group A), there were 237 upregulated proteins and 251 downregulated proteins. There were 145 upregulated and 204 downregulated proteins when comparing cells treated with oxaliplatin alone (group B) and cells treated with a combination of oxaliplatin and hyperthermia (group D) (Figure 2, Table 2).
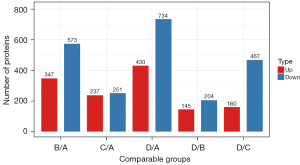
Table 2
| Compared sample name | Upregulated | Downregulated |
|---|---|---|
| B/A | 347 | 573 |
| C/A | 237 | 251 |
| D/A | 430 | 734 |
| D/B | 145 | 204 |
| D/C | 160 | 467 |
A, control cells; B, cells treated with oxaliplatin alone; C, cells treated with hyperthermia; D, cells treated with oxaliplatin and hyperthermia.
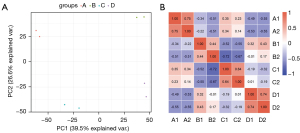
Samples repeatability inspection
Since each treatment was performed in duplicate, statistical analysis was performed to evaluate protein quantification repeatability. Figure 3 shows the results of the 2 duplicate samples from each of the 4 treatment groups. The Pearson correlation analysis heat map revealed that the Pearson index between the repeated groups is close to 1, indicating that the results between the repeated groups are positively correlated. The results of the 2 sets of graphs demonstrated that the reproducibility of the MS results is good and that the results are credible.
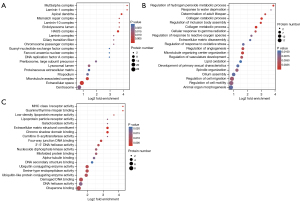
GO enrichment analysis
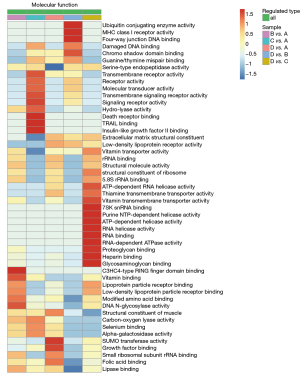
GO defines the concepts related to gene functions and how these functions are related to each other. GO describes the function from three fields, including cell composition, biological process, and molecular function. To determine the mechanisms by which hyperthermia enhances the sensitivity of ovarian cancer to chemotherapeutic drugs, the chemotherapy and hyperthermia combination group (group D) was compared with the chemotherapy-only group (group B). Cell composition analysis showed that the differentially expressed proteins between groups B and D are located in the extracellular space and the microtubule-related complexes (Figure 4A). The biological process analysis revealed that the formation of the microtubule tissue center and the differential proteins related to the regulation of angiogenesis were obviously aggregated (Figure 4B). In terms of molecular functions, the functions of most differential proteins were enriched in the binding of damaged DNA (Figure 4C).
Cluster analysis
Cluster analysis was performed to identify the correlation between the differentially expressed proteins in the different treatment groups. The cluster analysis related to molecular function demonstrated that the differentially expressed proteins between cells treated with a combination of chemotherapy and hyperthermia (group D) and cells treated with chemotherapy alone (group B) were enriched in ubiquitin conjugating enzyme activity, four-way junction DNA binding, and damaged DNA binding (Figure 5). This suggested that the mechanism by which hyperthermia enhances the sensitivity of ovarian cancer cells to chemotherapy is related to the repair of DNA damage. Therefore, it appears that thermochemotherapy regulates the synthesis of related proteins at the level of DNA damage repair and affects the repair of damaged DNA, resulting in unstable cell genome and cell death.
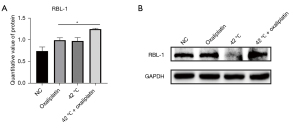
Hyperthermia combined with chemotherapy increases retinoblastoma-like protein 1 (RBL1/p107) expression in ovarian cancer cells
From the MS results, the top 20 differentially expressed proteins were selected for further analysis. There were 10 upregulated proteins, including CLU, RBL1, ANLN, COX17, RIOK1, KBTBD2, RAD51, UBE2T, SNIP1, and AK4; and 10 downregulated proteins, including HPF1, SMCHD1, LRBA, CHD3, TTC19, RNGTT, ASCC3, MSH6, CHD1L, and POLE2 (Tables 3,4). Among these proteins, more than two-thirds are involved in the DNA damage repair process. To confirm the MS results, the expression of the RBL-1/p107 protein was examined using Western blot. Indeed, consistent with the MS results (Figure 6A), RBL-1/p107 protein expression was significantly increased in cells that received thermochemotherapy combination treatment compared to cells treated with chemotherapy or hyperthermia alone (Figure 6B). The Rb family proteins (pRb/p105, Rb2/p130, and RBL1/p107) participate in transcription repression and tumor suppression, and can affect cell cycle control (17,18). Studies have demonstrated that pRb, pRb2/p130, and p107 can inhibit E2F responsive promoters, block gene transcription and cell cycle progression, and initiate apoptotic processes and differentiation by interacting with different E2F family factors (19,20). Recently, reports have suggested that the Rb gene family is important for maintaining genomic integrity, such as protecting cells from double-strand breaks (DSBs) that arise after DNA damage or during DNA replication (16).
Table 3
| Protein accession | Gene name | Protein description | D/B ratio |
|---|---|---|---|
| P10909 | CLU | Clusterin | 2.544 |
| P28749 | RBL1 | Retinoblastoma-like protein 1 | 1.2605 |
| Q9NQW6 | ANLN | Anillin | 1.2275 |
| Q14061 | COX17 | Cytochrome c oxidase copper chaperone | 1.2205 |
| Q9BRS2 | RIOK1 | Serine/threonine-protein kinase RIO1 | 1.2165 |
| Q8IY47 | KBTBD2 | Kelch repeat and BTB domain-containing protein 2 | 1.2125 |
| Q06609 | RAD51 | DNA repair protein RAD51 homolog 1 | 1.21 |
| Q9NPD8 | UBE2T | Ubiquitin-conjugating enzyme E2 T | 1.208 |
| Q8TAD8 | SNIP1 | Smad nuclear-interacting protein 1 | 1.206 |
| P27144 | AK4 | Adenylate kinase 4, mitochondrial | 1.2045 |
D/B ratio >1.2 is regarded as the change threshold of significant upregulation.
Table 4
| Protein accession | Gene name | Protein description | D/B ratio |
|---|---|---|---|
| Q9NWY4 | HPF1 | Histone PARylation factor 1 | 0.8315 |
| A6NHR9 | SMCHD1 | Structural maintenance of chromosomes flexible hinge domain-containing protein 1 | 0.831 |
| P50851 | LRBA | Lipopolysaccharide-responsive and beige-like anchor protein | 0.8215 |
| Q12873 | CHD3 | Chromodomain-helicase-DNA-binding protein 3 | 0.821 |
| Q6DKK2 | TTC19 | Tetratricopeptide repeat protein 19, mitochondrial | 0.8205 |
| O60942 | RNGTT | mRNA-capping enzyme | 0.8115 |
| Q8N3C0 | ASCC3 | Activating signal cointegrator 1 complex subunit 3 | 0.778 |
| P52701 | MSH6 | DNA mismatch repair protein Msh6 | 0.7595 |
| Q86WJ1 | CHD1L | Chromodomain-helicase-DNA-binding protein 1-like | 0.687 |
| P56282 | POLE2 | DNA polymerase epsilon subunit 2 | 0.6235 |
D/B ratio <1/1.2 is regarded as the change threshold of significant downregulation (CV-value <0.1).
Discussion
There is much clinical experimental data demonstrating that the survival of patients with ovarian cancer can be significantly prolonged by HIPEC (21). Indeed, the synergistic mechanisms of hyperthermia and chemotherapy is the focus of much research related to ovarian cancer treatment options (22). The development of proteomics based on LC-MS has facilitated the understanding of cellular and physiological processes involved in diseases, and the identification of novel biomarkers (14,23). Through massive proteomics analyses, there is now comprehensive information regarding protein regulation related to thermochemotherapy. In the current study, MS data analysis revealed that the differentially expressed proteins in thermochemotherapy-treated cells were mainly involved in ubiquitin conjugating enzyme activity, four-way junction DNA binding, and damaged DNA binding (Figure 5). It is well known that ubiquitin-conjugating enzymes promote DNA damage response (DDR) by integrating DNA repair and cell cycle checkpoint activation (24,25). Four-way junction DNA binding and damaged DNA binding in biology are involved in DNA damage repair (26). This suggested that hyperthermia mainly affects the DNA damage repair process in heat-sensitive ovarian cancer cells to enhance their sensitivity to chemotherapeutic drugs. The top 20 differentially expressed proteins were selected for further analysis. Indeed, two-thirds these proteins were involved in DNA damage repair. Western blot analysis confirmed that the protein expression of RBL1/P107 was significantly increased after thermochemotherapy, which was consistent with the MS results (Figure 6). Therefore, the DNA repair gene represented by RBL1/P107 may be an important target for hyperthermia to enhance the sensitivity of heat-sensitive ovarian cancer cells to chemotherapy. The retinoblastoma tumor (RB) suppressor is important in promoting cell cycle progression (27,28) and regulating chromatin functions to maintain genomic stability, including participating in DNA repair, condensing chromosome, telomere maintenance, and silencing of repetitive regions (29). However, the function of RBL1/p107 in DNA damage repair remains unclear, and there is currently a paucity of data reporting its association with hyperthermia. The latest research shows that hyperthermia inhibits cellular glycolysis, but in heat-resistant cells, their mitochondrial membrane potential is activated, which can compensate for the loss of ATP production due to the inhibition of glycolysis by hyperthermia (30). However, the mechanism of heat-resistant cells at the level of DNA damage repair still needs to be further explored. Our future investigations will include in vitro knockdown and overexpression experiments to identify the key DNA damage repair signaling pathways related to RBL1/p107, as well as characterization experiments in some ovarian cancer cells following thermochemotherapy. The findings will be verified using primary cells extracted from human tumor samples. Further research will focus on developing targeted small molecular drugs that simulate heat treatment. The efficacy of such agents at improving the sensitivity of heat-resistant cells to chemotherapeutics will be assessed.
Conclusions
This quantitative proteomics and MS study provided strong evidence that hyperthermia increases the sensitivity of tumor cells to chemotherapeutics through interfering with DNA damage repair, possibly through targeting the RBL1/p107 protein.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (Grant Nos. 81772773, 81302275, U1967220, and 12075165) and the Suzhou Clinical Key Technology Project (No. LCZX201705).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-955/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-955/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-955/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kurpeshev OK, Florovskaya NY, Lebedeva TV. Results of palliative thermochemotherapy for colorectal cancer metastases to the liver. Vopr Onkol 2016;62:85-90. [PubMed]
- Mi D, Li Z, Yang K, et al. Thermo-chemotherapy of GP or TP for advanced non-small cell lung cancer: a systematic review. Zhongguo Fei Ai Za Zhi 2012;15:456-64. [PubMed]
- Colombo R. Combined treatment with local thermo-chemotherapy for non muscle invasive bladder cancer. The present role in the light of acquired data and preliminary cumulative clinical experiences. Arch Ital Urol Androl 2008;80:149-56. [PubMed]
- Kleef R, Kekic S, Ludwig N. Successful treatment of advanced ovarian cancer with thermochemotherapy and adjuvant immune therapy. Case Rep Oncol 2012;5:212-5. [Crossref] [PubMed]
- Spiliotis J. Hyperthermic intraperitoneal chemotherapy in ovarian cancer: Qui Bono? Ann Transl Med 2020;8:1708. [Crossref] [PubMed]
- Di Giorgio A, De Iaco P, De Simone M, et al. Cytoreduction (Peritonectomy Procedures) Combined with Hyperthermic Intraperitoneal Chemotherapy (HIPEC) in Advanced Ovarian Cancer: Retrospective Italian Multicenter Observational Study of 511 Cases. Ann Surg Oncol 2017;24:914-22. [Crossref] [PubMed]
- Sukovas A, Silkuniene G, Trumbeckaite S, et al. Hyperthermia potentiates cisplatin cytotoxicity and negative effects on mitochondrial functions in OVCAR-3 cells. J Bioenerg Biomembr 2019;51:301-10. [Crossref] [PubMed]
- Hildebrandt B, Wust P, Ahlers O, et al. The cellular and molecular basis of hyperthermia. Crit Rev Oncol Hematol 2002;43:33-56. [Crossref] [PubMed]
- Hatakeyama H, Wu SY, Lyons YA, et al. Role of CTGF in Sensitivity to Hyperthermia in Ovarian and Uterine Cancers. Cell Rep 2016;17:1621-31. [Crossref] [PubMed]
- Oei AL, Kok HP, Oei SB, et al. Molecular and biological rationale of hyperthermia as radio- and chemosensitizer. Adv Drug Deliv Rev 2020;163-164:84-97. [Crossref] [PubMed]
- Luzhin AV, Avanesyan B, Velichko AK, et al. Chromatin Trapping of Factors Involved in DNA Replication and Repair Underlies Heat-Induced Radio- and Chemosensitization. Cells 2020;9:1423. [Crossref] [PubMed]
- Xie H, Huang H, Tang M, et al. iTRAQ-Based Quantitative Proteomics Suggests Synaptic Mitochondrial Dysfunction in the Hippocampus of Rats Susceptible to Chronic Mild Stress. Neurochem Res 2018;43:2372-83. [Crossref] [PubMed]
- Liu X, Zheng W, Wang W, et al. A new panel of pancreatic cancer biomarkers discovered using a mass spectrometry-based pipeline. Br J Cancer 2017;117:1846-54. [Crossref] [PubMed]
- Keshishian H, Burgess MW, Gillette MA, et al. Multiplexed, Quantitative Workflow for Sensitive Biomarker Discovery in Plasma Yields Novel Candidates for Early Myocardial Injury. Mol Cell Proteomics 2015;14:2375-93. [Crossref] [PubMed]
- Henningsen K, Palmfeldt J, Christiansen S, et al. Candidate hippocampal biomarkers of susceptibility and resilience to stress in a rat model of depression. Mol Cell Proteomics 2012;11:M111.016428.
- Genovese C, Trani D, Caputi M, et al. Cell cycle control and beyond: emerging roles for the retinoblastoma gene family. Oncogene 2006;25:5201-9. [Crossref] [PubMed]
- Claudio PP, Howard CM, Baldi A, et al. p130/pRb2 has growth suppressive properties similar to yet distinctive from those of retinoblastoma family members pRb and p107. Cancer Res 1994;54:5556-60. [PubMed]
- Dimova DK, Dyson NJ. The E2F transcriptional network: old acquaintances with new faces. Oncogene 2005;24:2810-26. [Crossref] [PubMed]
- Brehm A, Miska EA, McCance DJ, et al. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 1998;391:597-601. [Crossref] [PubMed]
- Magnaghi-Jaulin L, Groisman R, Naguibneva I, et al. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 1998;391:601-5. [Crossref] [PubMed]
- Tsuyoshi H, Inoue D, Kurokawa T, et al. Hyperthermic intraperitoneal chemotherapy (HIPEC) for gynecological cancer. J Obstet Gynaecol Res 2020;46:1661-71. [Crossref] [PubMed]
- Sugarbaker PH, Van der Speeten K. Surgical technology and pharmacology of hyperthermic perioperative chemotherapy. J Gastrointest Oncol 2016;7:29-44. [PubMed]
- Chen C, Jiang X, Li Y, et al. Low-dose oral copper treatment changes the hippocampal phosphoproteomic profile and perturbs mitochondrial function in a mouse model of Alzheimer's disease. Free Radic Biol Med 2019;135:144-56. [Crossref] [PubMed]
- Sun J, Zhu Z, Li W, et al. UBE2T-regulated H2AX monoubiquitination induces hepatocellular carcinoma radioresistance by facilitating CHK1 activation. J Exp Clin Cancer Res 2020;39:222. [Crossref] [PubMed]
- Bui QT, Hong JH, Kwak M, et al. Ubiquitin-Conjugating Enzymes in Cancer. Cells 2021;10:1383. [Crossref] [PubMed]
- van Rixel VHS, Busemann A, Wissingh MF, et al. Induction of a Four-Way Junction Structure in the DNA Palindromic Hexanucleotide 5'-d(CGTACG)-3' by a Mononuclear Platinum Complex. Angew Chem Int Ed Engl 2019;58:9378-82. [Crossref] [PubMed]
- Dick FA, Rubin SM. Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol 2013;14:297-306. [Crossref] [PubMed]
- van den Heuvel S, Dyson NJ. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol 2008;9:713-24. [Crossref] [PubMed]
- Vélez-Cruz R, Johnson DG. The Retinoblastoma (RB) Tumor Suppressor: Pushing Back against Genome Instability on Multiple Fronts. Int J Mol Sci 2017;18:1776. [Crossref] [PubMed]
- Kanamori T, Miyazaki N, Aoki S, et al. Investigation of energy metabolic dynamism in hyperthermia-resistant ovarian and uterine cancer cells under heat stress. Sci Rep 2021;11:14726. [Crossref] [PubMed]


