
Clostridium butyricum inhibits the progression of colorectal cancer and alleviates intestinal inflammation via the myeloid differentiation factor 88 (MyD88)-nuclear factor-kappa B (NF-κB) signaling pathway
Introduction
Colorectal cancer (CRC), a high-incidence gastrointestinal cancer, is the world's second highest cause of cancer mortality (1). Aside from hereditary CRC cases, which account for 5% of all CRC cases, the majority of CRC cases develop sporadically as a result of complicated risk factors such as aging, gender, obesity, and other lifestyle variables (2). Inflammatory bowel disease (IBD), which includes ulcerative colitis (UC) and Crohn’s disease (CD), is a major risk factor for CRC, and is also known as IBD-associated or colitis-associated CRC (CA-CRC) (3).
The gastrointestinal tract is the primary habitat for host microbiota, which is indispensable for health. The healthy status of gut microbiota has a favorable influence on disease prevention by resisting gut pathogens, reducing inflammation, and influencing the immune system (4). On the contrary, the dysbiosis of gut microbiota has detrimental effects on the aggravation of low-grade inflammation, the stimulation of immune responses, and the occurrence of CRC (5). The interactions between host factors and gut microbiota are crucial to the initiation and progression of CA-CRC elicited by chronic inflammation and other human diseases, which showed the importance of Clostridium butyricum (CB) on medical science for therapeutic purposes (6). Given the intricacies of CA-CRC induction, dissecting the mechanisms and understanding a path to improved treatment efficacy is required.
Considering its significance, the intervention of gut microbiota paves the way for the treatment of CA-CRC. CB, a spore-forming and short-chain fatty acid (SCFA)-producing probiotic, has long been utilized in the treatment of diarrhea (7), neurodegenerative diseases (8), and has been shown to ameliorate Parkinson’s disease in mice (9). Butyrate is the dominate products produced by CB, which has been found important effects on regulating intestinal immune system (8), modulating intestinal metabolite homeostasis (10) and inhibiting the proliferation of CRC cell (11). According to growing research, CB keeps the tight connections of the intestines intact (12), modulates the microbiota, and augments the abundance of probiotics (13). It was also found that CB protects cells from oxidative damage(14). Recently, a study demonstrated that CB inhibited the progression of CRC in high-fat diet APCmin/+ mice, potentially through inhibiting the Wnt/beta-catenin signaling pathway (15). Not merely having an influence on the development of CRC, CB also has promising supplementary effects on immune checkpoint blockade (ICB) therapy of lung cancer (16). Although the effects of CB have been studied in part, the mechanisms of CB on CA-CRC are not clear.
MyD88, a universal regulatory factor of innate immunity and inflammation, has been discovered to have a vital impact on CRC and the prognosis prediction of CRC (17). The activation of MyD88 leads to the activation of the nuclear factor-kappa B (NF-κB) signaling pathway and then gives rise to multiple downstream signals, which suppress apoptosis and mediate inflammation and autoimmune diseases (18). Overall, the abnormal activation of the MyD88-NF-κB signaling pathway promotes the development of CRC. Herein, we hypothesized whether CB might inhibit the progression of CA-CRC through modulating MyD88 and NF-κB and then conducted a series of experiments to verify our hypothesis.
In terms of the histopathological features, IBD, which is a risk factor for the progression of colitis-associated to CA-CRC, is similar to the AOM/DSS model mice. Herein, we established AOM/DSS model mice to study the impact of CB on CRC in vivo. In this study, we conducted animal experiments with CB treatment after the occurrence of CRC and explored the effect of CB on CRC cell lines in vitro comprehensively. We also studied the potential mechanisms by which CB functions. Our study will highlight the theoretical basis of their correlation and strengthen the feasibility that CB assists in the treatment of CRC. We present the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1670/rc).
Methods
Bacterial strains and culture conditions
CB (ATCC 19398) was purchased from Shanghai Fuxiang Biological Technology Co., Ltd. and cultured overnight at 37 ℃ under anaerobic conditions in brain heart infusion (BHI) broth. The bacteria were harvested by centrifugation (3,500 rpm × 10 minutes) and resuspended in sterile phosphate buffered saline (PBS) for animal and cell experiments. The supernatant was filtered with 0.22 µm pore size filters.
Animal experiments
Male 6-week-old C57BL/6J mice (about 16 g) obtained from Beijing HFK Bioscience Co., Ltd. were maintained in a specific pathogen-free (SPF) environment. All the mice (n=12) were acclimatized for 7 days before the animal experiments and then received a single dose of azoxymethane (AOM) 12.5 mg/kg per mouse by intraperitoneal injection. All the mice started 3 cycles of dextran sulfate sodium salt (DSS) treatment. For the first cycle of DSS, mice received drinking water with 3% DSS (w/v) for 5 days followed by 14 days of normal drinking water. For the next 2 cycles of DSS, mice received drinking water with 2.5% DSS (w/v) for 5 days followed by 14 days of normal drinking water. At the 41st day (the end of 5 days 2.5% DSS water drinking followed by 7 days of normal water drinking of the second DSS cycle), 10 mice were randomly divided into 2 groups [control (Ctrl) and CB group] with the same mean average body weight and 5 mice in each group. Besides the establishment of the normal CRC model, the mice in the CB group were administered CB [1×108 colony forming unit (CFU) in 100 µL PBS] by gavage gently 3 times a week until the end of animal experiments. The mice in the Ctrl group received an equivalent volume and frequency of PBS. All of the animals accepted mercy killing through cervical dislocation when rectal prolapse was observed in 50% of the animals. Tumor volume was calculated according to the formula (A × B2)/2, in which A and B are the long and short dimensions, respectively. A protocol was prepared before the study without registration. Animal experiments were performed under a project license (No. NCC2021A290) granted by institutional ethics committee of Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, in compliance with Chinese national guidelines for the care and use of animals.
Cell culture and treatment
Human CRC cell lines (HT-29 and CT26) were obtained from the National Infrastructure of Cell Line Resource (Beijing, China) and cultured in RPMI-1640 medium with 10% (v/v) fetal bovine serum (FBS) and 1% (v/v) penicillin-streptomycin liquid. Cells were incubated in a humidified incubator at 37 ℃ with 5% CO2. Cells were cultured in 6-well plates until 70–80% confluency in RPMI-1640 medium with 10% FBS and then co-cultured with different multiplicities of infection (MOIs) of CB for 24 hours to extract total RNA and protein. Each MOI set 3 duplicated wells.
Enzyme linked immunosorbent assay (ELISA)
After the collection of blood with anticoagulation, the samples were centrifuged for 15 minutes at 1,000 ×g at 2–8 ℃ within 30 minutes and stored in aliquot at −80 ℃ for later use. ELISA kits were used to test IL-10 (Cloud-Clone Corp, Wuhan) and IL-6 (4A Biotech, Beijing) in accordance with the manufacturer’s protocol. After bringing samples to room temperature, 100 µL each of dilutions of standard (read Reagent Preparation), blank and samples were added into the appropriate wells and incubated for 1 hour at 37 ℃ covered with the plate sealer, then remove the liquid of each well. 100 µL primary antibodies working solution was added to each well and incubated 1 hour at 37 ℃. Then 100 µL secondary antibody working solution was added to each well and incubated 1 hour at 37 ℃. Wash the well with 350 µL wash solution 3 times at the end of incubation of each antibody. Add 90 µL of Substrate Solution to each well. Cover with a new Plate sealer and incubate for 10–20 minutes protecting from the light. Then add 50 µL of Stop Solution to each well. The liquid will turn yellow from blue by the addition of Stop solution. At last, the plates were conducted measurement at 490 nm with a universal microplate reader (Bio-Tek, USA).
Histopathological observation and terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assays
Colons and tumors were collected and fixed with 4% (w/v) paraformaldehyde, dehydrated, and paraffin-embedded for sectioning and hematoxylin-eosin staining. The apoptosis of tumor cells in tumor tissues was detected by a TUNEL assay. Sections of paraffin-embedded tissue were deparaffinized and rehydrated with xylene and gradient alcohol, respectively. Then, the sections were treated in accordance with the manufacturer's protocol (Beyotime). After being dewaxed with xylene and rehydrated with gradient alcohol, the sections of paraffin-embedded tissue were digested with 20 µg/mL proteinase K for 20 minutes followed by washing three times with PBS. Then add mixture of Terminal Deoxyribonucleotidyl Transferase (TdT) and dUTP labeled fluorescein isothiocyanate (FITC) to the tissue on the sections for 1 hour at 37 ℃ protecting from the light. At last, the sections were counterstained with 4',6-diamidine-2-phenylindole dihydrochloride (DAPI)-antifade solution. Fluorescent images were captured with a confocal laser scanning microscope (CLSM) (Nikon, Japan). The numbers of apoptotic and total cells were counted by Image J (v1.8.0), and then the percentages of apoptotic cells were calculated.
Gut microbiota analysis through 16s rDNA sequencing
A total of 15 fecal samples were collected for 16S rRNA gene sequencing (Majorbio, Shanghai). Among these samples, 5 samples (original group) were collected from 5 mice in the same mouse cage while grouping and stored at −80 ℃. After euthanasia, the intestinal contents in mice were collected in a sterile environment and stored at −80 ℃. The Illumina MiSeq was used to sequence the V1–V2 hypervariable regions of microbiota 16S rRNA genes. Microbiota was analyzed including operational taxonomic unit (OTU), alpha diversity, community analysis, and beta diversity.
Fluorescence in situ hybridization (FISH) assays
FISH was used to determine the location and enrichment of CB. After being dewaxed with xylene and rehydrated with gradient alcohol, the sections of paraffin-embedded tissue were treated with 90 ℃ deionized water. The tissue was then digested for 15–30 minutes at 37 ℃ with 1 mg/mL pepsin. CB FISH probe (Future Biotech, Beijing) labeled with FITC and bacterial universal probe (Future Biotech, Beijing) labeled with CY5 were used for hybridization for 5 minutes at 80 ℃ under the conditions of protection from light, followed by 16 hours at 37 ℃. Lastly, sections were washed twice with 0.1% NP40 (Nonidet P40)/2 × SSC (saline sodium citrate) then 70% alcohol (v/v) and counterstained with DAPI-antifade solution. Fluorescent images were captured with a PE double spinning disk confocal (PerkinElmer, USA). The intensity of the fluorescent signal was assessed by Image J (v1.8.0).
Cell proliferation assay
Cell proliferation was assessed by the MTT colorimetric assay. The cells were seeded in 96-well plates at a density of 1,500–2,000 cells/well with 150 µL culture medium and treated with different MOIs of CB, different concentrations of sodium butyrate (NaB) (Solarbio, Beijing), and CB supernatant. Then, 5–10 µL of MTT was added into the cell plates at specific time points and cells were incubated for 2–4 hours per day at 37 ℃. Finally, intracellular purple formazan was solubilized by DMSO and then quantified at 490 nm with a universal microplate reader (Bio-Tek, USA).
Real time quantitative PCR (RT-qPCR)
The total RNA of tissues and cells was extracted using Trizol reagent and the RNApure Tissue and Cell Kit (Cowin Bio, USA), respectively, according to the manufacturer’s instructions. A total of 1 µg of total RNA was reverse transcribed using the PrimeScript RT Reagent Kit with gDNA Eraser (Perfect Real Time; Takara, Japan) to detect relative mRNAs. Primer sequences used in our study were showed in Table 1. Quantitative real-time PCR was performed with Roter Gene Q (QIAGEN, Germany) using specific primers, and the real-time PCR mixture (TB Green® Premix Ex TaqTM, Takara, Japan) was prepared according to the manufacturer’s instructions. Raw data was analyzed with QuantStudio TM Real-Time PCR Software (version 1.1) and fold change was calculated by the 2−ΔΔCt method. Beta actin served as an internal reference gene.
Table 1
| Gene name | Species | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|---|
| MyD88 | Human | CCGCCTGTCTCTGTTCTTG | GTCCGCTTGTGTCTCCAGTT |
| MyD88 | Mouse | ATCGCTGTTCTTGAACCCTCG | CTCACGGTCTAACAAGGCCAG |
| RelA/p65 | Human | ATGTGGAGATCATTGAGCAGC | CCTGGTCCTGTGTAGCCATT |
| RelA/p65 | Mouse | ACTGCCGGGATGGCTACTAT | TCTGGATTCGCTGGCTAATGG |
| Beta-actin | Human | TCACCCACACTGTGCCCATCTACGA | CAGCGGAACCGCTCATTGCCAATGG |
| Beta-actin | Mouse | GTGACGTTGACATCCGTAAAGA | GCCGGACTCATCGTACTCC |
Western blotting
Total protein was extracted from tissues and cells using highly efficient RIPA lysis buffer and quantified using the BCA Protein Assay Kit (Pierce Inc., USA). A total of 35 µg protein was electrophoresed through SDS-PAGE and transferred to PVDF membranes (Millipore, USA). After blocking for 0.5 hours with Ncm Blot blocking buffer (NCM Biotech, China), the membranes were incubated with primary antibodies including β-actin (42 KDa) (Proteintech, Wuhan), MyD88 (33 KDa) (Proteintech, Wuhan), and p65 (65 KDa) (Abcam, Shanghai) at 4 ℃ overnight. Membranes were then incubated with secondary antibodies labeled with HRP (Proteintech, Wuhan) for 1.5 hours successively. Finally, signals were detected using ImmobilonTM Western Chemiluminescent HRP Substrate (Millipore, USA). The images were analyzed by ImageJ (v1.8.0). Beta actin served as an internal reference protein.
Immunohistochemistry (IHC)
Slides of colon tissues after pretreatment (dewaxing, rehydration, and antigen retrieval) were blocked by 3% (m/v) H2O2 for 30 minutes and 5% (m/v) skim milk for 4 hours. Then, the slides were incubated with anti-Ki-67 antibody (Proteintech, Wuhan) at 4 ℃ overnight and secondary antibodies labeled with HRP (Proteintech, Wuhan) for 0.5 hours successively. After washing with PBS, fresh DAB chromogenic fluid (Solarbio, Beijing) was added to the tissues on the slides for approximately 1 minute and sections were observed under a microscope to control the color. After restaining the nucleus, the slides were dehydrated in gradient alcohol and sealed with resinene. The percentage of Ki-67-positive cells was analyzed by Image J (v1.8.0).
Statistical analysis
The Student’s t test was used to compare the means of 2 independent groups by SPSS. Statistical images were generated using GraphPad Prism 8.4.2. All statistical tests were two-sided. P<0.05 was considered statistically significant. All measurements and statistical analyses were carried out by another person in order to control for bias.
Results
CB inhibited the development of CRC in vivo
The AOM/DSS model diagram is shown in Figure 1A. Two mice received euthanasia to identify the occurrence of colon tumors (Figure 1B). Then, mice were divided into 2 groups (Ctrl and CB group), and mice in the CB group were treated with CB for 40 days during the establishment of the AOM/DSS model. The occurrence of colon tumors was assessed at the end of the animal experiments. Representative images of the colon in the Ctrl and CB groups are shown in Figure 1C. The majority of tumors formed in the distal colon and rectum, and partly clustered on 1 or 2 locations. The number and volume per mouse were quantified and shown in Figure 1D and Figure 1E, respectively. The Ctrl and CB groups had an average of 4.200 (±0.837) and 2.400 (±0.894) tumors per mouse, respectively (P<0.05). The average volume of tumors per mouse in the Ctrl and CB groups was 5 mm3 (±5.079) and 2.469 mm3 (±4.121), respectively (P<0.05). The histological characteristics showed that more tumors were present in the Ctrl group and most of the tumors in the CB group were latent (Figure 1F). The results indicated that CB suppressed the progression of tumors.
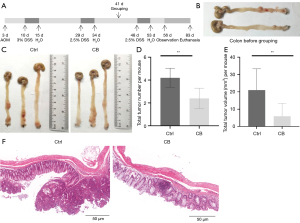
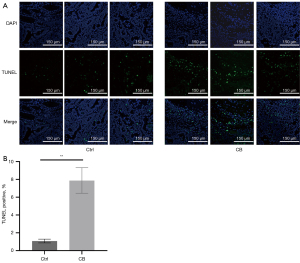
We also conducted TUNEL staining assays of the tumor tissues to determine the impact of CB on cancer cell apoptosis. Observation under confocal microscopy (Figure 2A) and statistical analysis (Figure 2B) of TUNEL staining showed that the number and percentage of apoptotic cells in the tumor tissues of mice significantly increased in the CB group compared with that in the Ctrl group (P<0.05), which showed the effect of CB on inducing the death of cancer cells.
CB improved intestinal inflammation in AOM/DSS mice
We recorded the body weight of mice every 3 days after grouping (Figure 3A). The body weight of mice decreased during the DSS cycle and recovered gradually when normal drinking was resumed. During animal experiments, mice in the CB group were heavier than mice in the Ctrl group. At the end of the animal experiments, the body weight of mice in the CB group exceeded that in the Ctrl group (P>0.05). The colon length shortened while the mice suffered from inflammation. The statistical analysis of the colon length showed no significant difference, but the mean of the colon length in the CB group was higher than that in the Ctrl group (Figure 3B). The histological characteristics showed more plasma cell infiltration and more severe colonic crypt disorder in the Ctrl group compared to that in the CB group (Figure 3C).
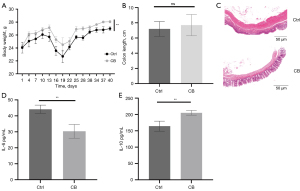
The levels of IL-6, an inflammatory cytokine, and IL-10, an anti-inflammatory cytokine, in plasma were quantified using ELISA (Figure 3D,3E). A higher concentration of IL-6 and lower concentration of IL-10 were found in mice of the Ctrl group compared with the CB group. These results demonstrated that administration of CB alleviated inflammation in mice.
CB rebuilt the composition of gut microbiota
The 16S rDNA sequencing of gut microbiota was performed to explore the role of CB in gut microbiota. With respect to the OTUs of bacteria, the OTU numbers in each group and shared OTUs are shown in Figure 4A. Besides most of the shared OTUs, a few specific OTUs existed in each group. The alpha diversity of gut microbiota was estimated by the Simpson index (Figure 4B). The Student’s t test for the Simpson index showed that CB changed the community diversity (Figure 4C).
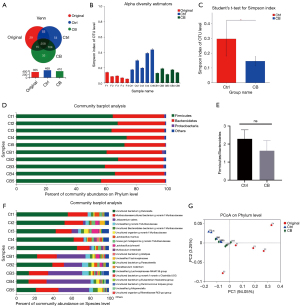
We compared the specific changes in bacterial abundance at the phylum level (Figure 4D). The average abundance of Bacteroidetes and Firmicutes in gut microbiota was, respectively, 30.6% and 68.28% in the Ctrl group and 39.19% and 59.20% in the CB group. We also calculated the Firmicutes/Bacteroidetes (F/B) ratio of the 2 groups. The average F/B ratio was 2.29 and 1.63 in the Ctrl and CB groups, respectively. This finding showed the effect of CB on increasing the F/B ratio to some extent, but with no significant difference in statistical analysis (Figure 4E). The analysis of bacterial abundance at the species level is shown in Figure 4F. It revealed more differences in bacterial abundance in the 2 groups.
The similarities and differences among samples in different groups were assessed using principal co-ordinates analysis (PCoA) of the beta diversity (Figure 4G). We were aware that the location of the samples in the Ctrl and CB groups showed no significant difference, but the location of samples in the original group was scattered, which suggested that DSS treatment influenced the beta diversity, and the response of mice to DSS showed a great difference.
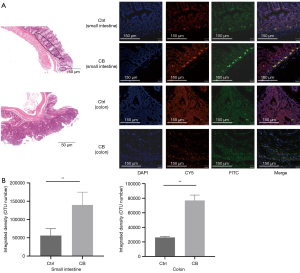
In order to determine the location and enrichment of CB in the intestine, we performed the FISH assay. Observation under confocal microscopy (Figure 5A) and statistical analysis (Figure 5B) of the fluorescence signal showed that CB in the CB group was prone to being enriched in the crypts of the small intestine and tumors of the colon. These findings demonstrated that CB could be enriched in the small intestine and infiltrate into the colon tumor.
CB restrained the proliferation of CRC cells and the expression of MyD88 and NF-κB/p65 in vitro
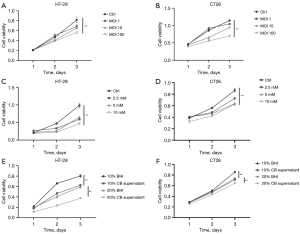
To identify the function of CB in the proliferation of CRC cell lines, we co-cultured the cell lines with CB and conducted the MTT assay. The proliferation of HT-29 cells (Figure 6A) and CT26 cells (Figure 6B) was inhibited by CB. Cell viability decreased progressively with the increase in the MOI of CB. We treated the CRC cell lines with different concentrations of NaB (Figure 6C,6D), a primary metabolite of CB, and CB supernatant (Figure 6E,6F) to further clarify that CB inhibited the proliferation of CRC cells not through bacterial pollution.
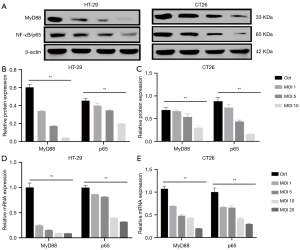
NF-κB, which is a transcription factor regulated by MyD88, plays an essential role in inflammation and the progression of CRC. To probe whether CB inhibited the progression of CRC and inflammation through restraining the expression of MyD88 and NF-κB, we explored the changes in the expression of MyD88 and NF-κB/p65 at the mRNA and protein levels. We observed the protein expression of MyD88 and NF-κB/p65 in HT-29 cells and CT26 cells (Figure 7A). The quantitative analysis showed that the relative MyD88 and p65 protein expression in HT-29 cells (Figure 7B) and CT26 cells (Figure 7C) was decreased with the increase in the MOI of CB. As shown in Figure 7D, the relative MyD88 and NF-κB/p65 mRNA expression in HT-29 cells was decreased with the increase of the MOI of CB, and the same trend was shown in CT26 cells (Figure 7E). These findings demonstrated that CB could inhibit the expression of MyD88 and NF-κB/p65, and when it comes to the expression of MyD88, HT-29 cells showed higher sensitivity to CB compared with CT26 cells.
CB restrained the proliferation of CRC cells and the expression of MyD88 and NF-κB/p65 in vivo
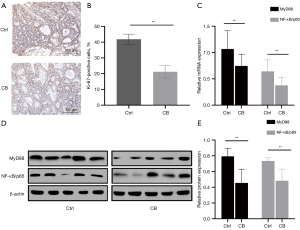
We conducted Ki-67 immunohistochemical staining to study the effects of CB on proliferation in the colon and colon tumors (Figure 8A). The statistical analysis showed a significant decrease in the percentage of Ki-67-positive cells (Figure 8B). Furthermore, we explored whether CB inhibited the expression of MyD88 and NF-κB/p65 in the colons of mice in the CB group compared with the Ctrl group. The results of real-time qPCR (Figure 8C) and western blot (Figure 8D,8E) showed a significant decrease in the CB group. Overall, CB inhibited the progression of CRC in vivo through suppressing the expression of MyD88 and NF-κB/p65.
Discussion
Our animal experiments showed that CB inhibited the formation of CRC significantly. The experiments conducted with CRC cell lines and tissues of AOM/DSS mice showed that CB restrained the proliferation of CRC and may influence the progression of CRC via MyD88-NF-κB signaling.
The gut microbiota, which is regarded as the second human genome, participates in food digestion, the integrity of intestinal mucosal barriers, and the maturation of immunological tissues (19). In a comprehensive investigation of CRC, the differences in gut microbiota between CRC patients and healthy people and the role of several bacteria, including Escherichia coli, Bacteriodes fragilis, and Fusobacterium nucleatum, in CRC carcinogenesis shed light on CRC treatment targeted at the gut microbiota and these special bacteria (20). CB as a safe probiotic has been found to have an important role in the regulation of gut microbiota (21) and inhibits intestinal tumor development in APCmin/+ mice (15). CB is reported to have the ability to protect the intestinal barrier through the activation of the Akt/mTOR signaling pathway (22). Research also revealed the mechanisms of CB in alleviating Enterotoxigenic Escherichia K88-induced oxidative damage (23). It is undeniable that CB has enormous potential in the treatment of CRC. However, all this existing research administered CB treatment to model animals from the beginning of the animal experiments, which only demonstrates that CB inhibits the formation of CRC. To investigate the effect of CB on the progression of CA-CRC, we treated AOM/DSS model mice with CB after the second DSS cycle for 40 days when the adenoma developed, so that we could explore the potential of CB for CRC treatment. We observed that both the tumor number and total volume per mouse decreased while the apoptosis of tumor cells increased after CB treatment. Thus, we conclude that CB inhibits the development of CA-CRC and induces the apoptosis of cancer cells in tumor tissue. When it comes to the detection of apoptosis, it would be better to add test results from other methods for a better study.
Chronic inflammation has been regarded as a characteristic of multiple cancers and plays an important role in the progression of most cancers (24). Probiotic supplements are a frequently-used way to modulate intestinal function (25). CB maintains intestinal homeostasis to suppress experimental acute pancreatitis (26) and also rescues virus-induced lung inflammation in mice (27). Furthermore, CB has long been known for its anti-inflammatory effect and mucosal barrier protection in intestinal disease (28), and it induces protect in D1 to ameliorate inflammation in antibiotic-induced intestinal disorder (29). IL-6 is synthesized in the initial stage of inflammation and moves through the bloodstream to cause a series of biological effects, including reducing the production of transferrin, fibronectin, and albumin and impairing the function of many organs (30). IL-10, an anti-inflammatory cytokine, is capable of restricting excessive inflammatory responses through the inhibition of pro-inflammatory cytokines and promoting tissue repair (31). Therefore, IL-6 and IL-10 are reasonable indicators for the evaluation of the inflammatory state. In this study, we estimated the inflammation of mice via recording the body weight, colon length, histopathological observations, and the levels of IL-6 and IL-10 in mice. Accordingly, we demonstrated that CB improves the state of inflammation in mice. Because of the lack of plasma samples, only two inflammatory indicators were measured in our study and the detection of more inflammatory indicators will be supportive.
The structure and composition of gut microbiota are some of the criteria used to estimate the state of the intestinal tract. Gut microbiota has close interactions with intestinal cells and is involved in many phases of CRC, including the initiation, progression, and treatment efficacy (20). A study in which germ-free mice received fecal samples from CRC patients reported that healthy mice treated with AOM (a commonly used mutagen) developed increased polyps and severe intestinal dysplasia after the gavage of stool samples from CRC patients (32). Much research has been performed on the relationship between inflammation and gut microbiota (33). Following inflammation, the diversity of gut microbiota and the abundance of probiotics decrease (13), while the F/B abundance ratio increases (34). On the basis of these changes in gut microbiota during inflammation, we analyzed the gut microbiota to determine the performance of CB. We demonstrated that the OTU number and alpha diversity of the Ctrl group was higher than the other 2 groups, while the F/B abundance ratio did not show a significant difference. The results are different from much of the previous research and can be attributed to the different treatment strategies in the animal experiments. Although there was only a slight difference in community abundance at the phylum level, we observed obvious differences at the species level. As for the beta diversity, the locations of samples in the Ctrl and CB groups gathered together and were distinct from the scattered location of the original group, which is caused by the different responses to DSS to some extent. In addition, we reported that CB is able to be enriched in the intestine and tumor, for which CB serves a function at specific location. Besides the effect of CB on the regulation of gut microbiota, it is also important to figure out the effects of changes in the gut microbiota during CB treatment on gene expression.
It has been found that MyD88-dependent signaling plays an important role in the mediation of microbiota dysbiosis and the malignant features of colitis (35). As a downstream effector of MyD88, NF-κB promotes tumor growth and survival, making it a potential target for anticancer therapy (36). It is also related to the angiogenesis and metastasis of tumors (37). A study reported that MyD88 promotes proliferation, migration, and invasion through the activation of the NF-κB/AP signaling pathway (38). Recent research found MyD88 and NF-κB activation in skeletal muscle atrophy (39) and TLR (Toll-like receptor) adaptor and MyD88 mutations could activate NF-κB expression to drive B cell lymphomagenesis (40). Verification of the effect of CB is always performed through in vivo animal experiments or cell lines treated with the culture medium of CB. We co-cultured CRC cell lines with CB to dissect the potential mechanisms. We demonstrated that CB inhibits the proliferation of CRC cells and the expression of MyD88 and NF-κB in cell lines and colon tissues. As for the different sensitivity of CRC cells to CB, we conducted an experiment with CRC cells that have higher survivability while co-culturing with CB. Therefore, CB orchestrates MyD88 and NF-κB to promote the progression of CRC. As for the interaction between cell receptors, such as TLRs, and MyD88, studies on the upstream of MyD88 is planned to conduct. The inhibitor of MyD88 is also on the list of subsequent studies to support the conclusion. This is also a limitation for this study.
Our study showed the effects of CB through different experimental designs, in which AOM/DSS mice were treated with CB at the beginning of CRC initiation, and the mechanisms were verified with CB instead of CB supernatant. All the results demonstrated the significant effects of CB on CRC and revealed the mechanisms of CB’s influence on CRC. Our study furthers the potential of CRC treatment using a CB supplement.
Acknowledgments
We would like to thank Chunxiao Gao (Peking University Fifth School of Clinical Medicine, Beijing, China; Center of Biotherapy, Beijing Hospital, National Center of Gerontology,Institute of Geriatric Medicine, Chinese Academy of Medical Sciences, Beijing, China) and Zhikai Han (State Key Laboratory of Molecular Oncology, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China) for technical support during the experiments.
Funding: This study was supported by the National Natural Science Foundation of China (Nos. 51972343, 51937011) and Research Projects of Cancer Hospital Chinese Academy of Medical Sciences (No. LC2020A24).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1670/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1670/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1670/coif). All authors report that this study was supported by the National Natural Science Foundation of China (Nos. 51972343, 51937011) and Research Projects of Cancer Hospital Chinese Academy of Medical Sciences (No. LC2020A24). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Animal experiments were performed under a project license (No. NCC2021A290) granted by institutional ethics committee of Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, in compliance with Chinese national guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Thrumurthy SG, Thrumurthy SS, Gilbert CE, et al. Colorectal adenocarcinoma: risks, prevention and diagnosis. BMJ 2016;354:i3590. [Crossref] [PubMed]
- Eaden JA, Abrams KR, Mayberry JF. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut 2001;48:526-35. [Crossref] [PubMed]
- Dos Reis SA, da Conceição LL, Siqueira NP, et al. Review of the mechanisms of probiotic actions in the prevention of colorectal cancer. Nutr Res 2017;37:1-19. [Crossref] [PubMed]
- Hills RD Jr, Pontefract BA, Mishcon HR, et al. Gut Microbiome: Profound Implications for Diet and Disease. Nutrients 2019;11:1613. [Crossref] [PubMed]
- Brennan CA, Garrett WS. Gut Microbiota, Inflammation, and Colorectal Cancer. Annu Rev Microbiol 2016;70:395-411. [Crossref] [PubMed]
- Seki H, Shiohara M, Matsumura T, et al. Prevention of antibiotic-associated diarrhea in children by Clostridium butyricum MIYAIRI. Pediatr Int 2003;45:86-90. [Crossref] [PubMed]
- Stoeva MK, Garcia-So J, Justice N, et al. Butyrate-producing human gut symbiont, Clostridium butyricum, and its role in health and disease. Gut Microbes 2021;13:1-28. [Crossref] [PubMed]
- Sun J, Li H, Jin Y, et al. Probiotic Clostridium butyricum ameliorated motor deficits in a mouse model of Parkinson's disease via gut microbiota-GLP-1 pathway. Brain Behav Immun 2021;91:703-15. [Crossref] [PubMed]
- Zhang L, Liu C, Jiang Q, et al. Butyrate in Energy Metabolism: There Is Still More to Learn. Trends Endocrinol Metab 2021;32:159-69. [Crossref] [PubMed]
- Li Q, Cao L, Tian Y, et al. Butyrate Suppresses the Proliferation of Colorectal Cancer Cells via Targeting Pyruvate Kinase M2 and Metabolic Reprogramming. Mol Cell Proteomics 2018;17:1531-45. [Crossref] [PubMed]
- Li H, Gong Y, Xie Y, et al. Clostridium butyricum protects the epithelial barrier by maintaining tight junction protein expression and regulating microflora in a murine model of dextran sodium sulfate-induced colitis. Scand J Gastroenterol 2018;53:1031-42. [Crossref] [PubMed]
- Hagihara M, Kuroki Y, Ariyoshi T, et al. Clostridium butyricum Modulates the Microbiome to Protect Intestinal Barrier Function in Mice with Antibiotic-Induced Dysbiosis. iScience 2020;23:100772. [Crossref] [PubMed]
- Dou C, Shang Z, Qiao J, et al. Clostridium butyricum Protects IPEC-J2 Cells from ETEC K88-Induced Oxidative Damage by Activating the Nrf2/ARE Signaling Pathway. Oxid Med Cell Longev 2021;2021:4464002. [Crossref] [PubMed]
- Chen D, Jin D, Huang S, et al. Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett 2020;469:456-67. [Crossref] [PubMed]
- Tomita Y, Ikeda T, Sakata S, et al. Association of Probiotic Clostridium butyricum Therapy with Survival and Response to Immune Checkpoint Blockade in Patients with Lung Cancer. Cancer Immunol Res 2020;8:1236-42. [Crossref] [PubMed]
- Li WL, Xiao MS, Zhang DF, et al. Mutation and expression analysis of the IDH1, IDH2, DNMT3A, and MYD88 genes in colorectal cancer. Gene 2014;546:263-70. [Crossref] [PubMed]
- Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-κB as the matchmaker. Nat Immunol 2011;12:715-23. [Crossref] [PubMed]
- Tang WH, Kitai T, Hazen SL. Gut Microbiota in Cardiovascular Health and Disease. Circ Res 2017;120:1183-96. [Crossref] [PubMed]
- Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol 2019;16:690-704. [Crossref] [PubMed]
- Liu M, Xie W, Wan X, et al. Clostridium butyricum modulates gut microbiota and reduces colitis associated colon cancer in mice. Int Immunopharmacol 2020;88:106862. [Crossref] [PubMed]
- Liu M, Xie W, Wan X, et al. Clostridium butyricum protects intestinal barrier function via upregulation of tight junction proteins and activation of the Akt/mTOR signaling pathway in a mouse model of dextran sodium sulfate-induced colitis. Exp Ther Med 2020;20:10. [Crossref] [PubMed]
- Li H, Shang Z, Liu X, et al. Clostridium butyricum Alleviates Enterotoxigenic Escherichia coli K88-Induced Oxidative Damage Through Regulating the p62-Keap1-Nrf2 Signaling Pathway and Remodeling the Cecal Microbial Community. Front Immunol 2021;12:771826. [Crossref] [PubMed]
- Taniguchi K, Karin M. NF-κB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol 2018;18:309-24. [Crossref] [PubMed]
- Guo Q, Goldenberg JZ, Humphrey C, et al. Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Database Syst Rev 2019;4:CD004827. [Crossref] [PubMed]
- Pan LL, Niu W, Fang X, et al. Clostridium butyricum Strains Suppress Experimental Acute Pancreatitis by Maintaining Intestinal Homeostasis. Mol Nutr Food Res 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Zhu W, Wang J, Zhao N, et al. Oral administration of Clostridium butyricum rescues streptomycin-exacerbated respiratory syncytial virus-induced lung inflammation in mice. Virulence 2021;12:2133-48. [Crossref] [PubMed]
- Li Y, Liu M, Liu H, et al. The Anti-Inflammatory Effect and Mucosal Barrier Protection of Clostridium butyricum RH2 in Ceftriaxone-Induced Intestinal Dysbacteriosis. Front Cell Infect Microbiol 2021;11:647048. [Crossref] [PubMed]
- Ariyoshi T, Hagihara M, Eguchi S, et al. Clostridium butyricum MIYAIRI 588-Induced Protectin D1 Has an Anti-inflammatory Effect on Antibiotic-Induced Intestinal Disorder. Front Microbiol 2020;11:587725. [Crossref] [PubMed]
- Tanaka T, Narazaki M, Kishimoto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb Perspect Biol 2014;6:a016295. [Crossref] [PubMed]
- Ouyang W, O'Garra A. IL-10 Family Cytokines IL-10 and IL-22: from Basic Science to Clinical Translation. Immunity 2019;50:871-91. [Crossref] [PubMed]
- Wong SH, Zhao L, Zhang X, et al. Gavage of Fecal Samples From Patients With Colorectal Cancer Promotes Intestinal Carcinogenesis in Germ-Free and Conventional Mice. Gastroenterology 2017;153:1621-1633.e6. [Crossref] [PubMed]
- Arthur JC, Perez-Chanona E, Mühlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012;338:120-3. [Crossref] [PubMed]
- Murphy EF, Cotter PD, Healy S, et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut 2010;59:1635-42. [Crossref] [PubMed]
- Wang L, Yu K, Zhang X, et al. Dual functional roles of the MyD88 signaling in colorectal cancer development. Biomed Pharmacother 2018;107:177-84. [Crossref] [PubMed]
- Perkins ND. The diverse and complex roles of NF-κB subunits in cancer. Nat Rev Cancer 2012;12:121-32. [Crossref] [PubMed]
- Patel M, Horgan PG, McMillan DC, et al. NF-κB pathways in the development and progression of colorectal cancer. Transl Res 2018;197:43-56. [Crossref] [PubMed]
- Zhu G, Cheng Z, Huang Y, et al. MyD88 mediates colorectal cancer cell proliferation, migration and invasion via NF-κB/AP-1 signaling pathway. Int J Mol Med 2020;45:131-40. [PubMed]
- Parveen A, Bohnert KR, Tomaz da Silva M, et al. MyD88-mediated signaling intercedes in neurogenic muscle atrophy through multiple mechanisms. FASEB J 2021;35:e21821. [Crossref] [PubMed]
- Cardona Gloria Y, Bernhart SH, Fillinger S, et al. Absence of Non-Canonical, Inhibitory MYD88 Splice Variants in B Cell Lymphomas Correlates With Sustained NF-κB Signaling. Front Immunol 2021;12:616451. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


