
Bromodomain-containing protein 4 activates cardiotrophin-like cytokine factor 1, an unfavorable prognostic biomarker, and promotes glioblastoma in vitro
Introduction
Glioblastoma (glioblastoma multiforme, GBM) is one of the most common and malignant grade IV glial tumors (1,2). Mutations (MUT) in isocitrate dehydrogenase (IDH) and O6-methylguanine-methyltransferase (MGMT) promoter methylation are 2 prognostic biomarkers of GBM (3). Typical molecular changes include MUT in receptor tyrosine kinase (RTK), rat sarcoma (RAS), phosphoinositide 3-kinase (PI3K), p53, and retinoblastoma protein (RB) signaling genes (4,5). Although surgery combined with radiation therapy and chemotherapy has been widely used for the treatment of GBM, the prognosis is still poor. Thus, it is necessary to identify all potential therapeutic targets for GBM and clarify their underlying mechanisms.
Increasing evidence indicates that immune cells and cytokines (6) in the tumor microenvironment are involved in GBM progression (7), prognosis (8), and therapy response (9). Cardiotrophin-like cytokine factor 1 (CLCF1) is a member of the IL-6 superfamily (10) and forms a heterodimer complex with cytokine receptor-like factor 1 (CRLF1). As a potent neurotrophic factor, CLCF1 competes with ciliary neurotrophic factor (CNTF) and binds ciliary neurotrophic factor receptor (CNTFR) (11). CLCF1 was also reported to bind and activate the ILST/gp130 receptor and Janus kinase (JAK)-STAT signaling cascade (12). Previous studies have shown that CLCF1 is involved in osteogenesis (13), osteoporosis (14), hematopoiesis (15), atherosclerosis (16,17), fibrosis (18), nerve regeneration (19), cold-induced sweating syndrome (20), inflammatory diseases (12), and lung cancer (11,16,21). CLCF1 was significant prognostic genes in both IDH-wild type (WT)/PTEN-MUT GBM and LGG patients, and was bond by the small molecule compound (+)-JQ1 as potential therapy for PTEN-mut glioma (22). However, further studies are needed to uncover the detailed function and regulation of CLCF1 in GBM.
Recently, the bioinformatics research and microarray technology has enabled us to understand the occurrence, development and metastasis of glioma at the molecular level. Bioinformatics research revealed that therapeutic molecular targets and the theoretical basis of tumor with systematic, accurate and effective way, which makes it possible to study the genetic changes and molecular mechanisms of glioma and provides a new idea for studying the molecular pathogenesis of various diseases. Bioinformatics research including gene ontology (GO), Kyoto encyclopedia of genes and genomes (KEGG), protein to protein interaction analysis (GSEA), etc. These methods help us identify the core driving genes of the disease and the abnormal regulatory pathways in glioblastoma. Gene Expression Profiling Interactive Analysis (GEPIA) database including the genes expression and the survival prognosis of glioblastoma patients.
Here, we first studied the expression changes and prognostic significance of CLCF1 in GBM based on the GEPIA database. Then, the function of CLCF1 in glioma cells was investigated with gene loss-of-function strategies. Next, the downstream target of CLCF1 in GBM was analyzed. Finally, we analyzed the transcriptional activators for high expression of CLCF1 in GBM tumor samples. This study deepens our understanding of CLCF1 in GBM and provides a potential target for GBM therapy. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1164/rc).
Methods
Differential expression and prognostic significance of CLCF1 in GBM datasets
The expression and prognostic significance of CLCF1 was investigated with The Cancer Genome Atlas (TCGA) GBM and Chinese Glioma Genome Atlas (CGGA) datasets using Gene Expression Profiling Interactive Analysis (GEPIA) (23). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture and transfection
The human GBM cell lines U87 and U251 were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences, Shanghai, China. U87 and U251 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Thermo Fisher Scientific, Waltham, MA, USA). Three shRNAs targeting CLCF1 were designed, and silencing efficiency was examined with real-time polymerase chain reaction (PCR).
The sequencing of shRNAs for CLCF1 were as follows: shRNA1 (5'-3'): GGCTGGGACCTATCTGAACTA; shRNA2 (5'-3'): GCTGGGACCTATCTGAACTAC; shRNA3 (5'-3'): GCGAAGCCTCAATGACAAACT. The primer sequences of CLCF1 and GAPDH (housekeeping gene) were as follows: CLCF1 F(5'-3'): TTTCAACGAGCCAGACTTCAAC, R(5'-3'): GAGGCCACGCAAGTAACACA; GAPDH F(5'-3'): GGAGCGAGATCCCTCCAAAAT, R(5'-3'): GGCTGTTGTCATACTTCTCATGG. Gene expression quantification was performed with the 2−ΔΔCT method in this study.
Cell proliferation assays
Cell proliferation was measured using the Cell Counting Kit 8 (CCK-8) assay. After 48 h of transfection, cells were seeded in 96-well plates at a density of 5×103 per well and cultured in an incubator for 0, 24, 48, and 72 h at 37 ℃. Then, 10 μL of CCK-8 solution (Beyotime Biotechnology, Shanghai, China) was added, and the cells were cultured in an incubator for 2 h. The OD450 was measured using a spectrophotometer (DS-11 FX; DeNovix, Wilmington, USA).
Cell cycle assay by flow cytometry
The effect of CLCF1 on cell cycle arrest in GBM cells was assessed using a flow cytometer. Cells were harvested, washed twice using phosphate-buffered saline (PBS), and fixed with 75% ethanol at 4 ℃ for at least 4 h. Then, the cells were collected and stained with PI solution/RNase A at 4 ℃ in the dark for 30 min, followed by flow cytometric analysis.
Apoptosis assay by flow cytometry
The effect of CLCF1 on apoptosis in GBM cells was assessed using a flow cytometer. U251 cells were transfected and cultured for 24 h. Then, the cells were digested, washed, and resuspended in PBS. Finally, the cells were stained using Annexin V-FITC at 4 ℃ in the dark for 20 min and analyzed by flow cytometry.
Wound healing assay
Take cells from each group and add them into the well plate. There are 5 horizontal lines and 5×105 cells. After overnight incubation, 200 μL gun head is perpendicular to the horizontal line scratch on the back, and wash away the scratched cells. After 48 hours of treatment, take samples and take photos.
Transwell assay
U251 cells were routinely digested, centrifuged and collected, and cultured with serum-free DMEM. The cells were resuspended in the medium and made into cells with a density of 2.5×104 cells/mL suspension. 200 mL cell suspension was inoculated into the upper chamber of transwell chamber, 800 mL DMEM medium containing 10% fetal bovine was added to the lower chamber. Incubate in 37 ℃ incubator for 48 h. Take out the transwell chamber, gently wipe the non-invasion cells in the upper chamber with a cotton swab, wash with PBS for 3 times, and dry them at room temperature for about 10 min. The cells were stained with 100 mL of crystal violet. Let it stand at room temperature for 20 min, and then wash it with PBS for 3 times. The invasion cells were observed and counted under the microscope. The experiment was repeated three times.
Gene set enrichment analysis (GSEA) of CLCF1-related pathways
GBM samples were separated into 2 groups (high and low) based on the median CLCF1 expression. GSEA was applied to identify the differences in the MSigDB (https://www.gsea-msigdb.org/gsea/msigdb/collections.jsp) C2 collection between the high group and low group.
Dual luciferase assay
HEK-293T cells were plated on a 12-well plate and cultured at 37 ℃ in a humidified incubator with 5% CO2. One day after culture, the cells in each well were co-transfected with pcDNA3.1. The cells were also co-transfected with pcDNA3.1-CLCF1-WT or pcDNA3.1-CLCF1-MUT using Renilla luciferase expression vectors. The bromodomain-containing protein 4 (BRD4) inhibitor JQ1 was added to the 293T cells. After 48 hours of culture, luciferase activity was detected according to the steps of double luciferase reporter gene detection kit.
Statistical analysis
The statistical analysis was conducted with GraphPad Prism 8.0. A t-test or one-way ANOVA was used to compare the difference between 2 or more groups. All cell experiments were replicated 3 times, and a P value <0.05 was considered statistically significant.
Results
CLCF1 is an unfavorable prognostic marker in GBM
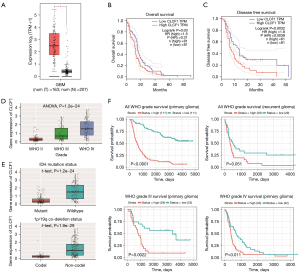
To explore the clinical significance of CLCF1 expression in GBM, we first analyzed the differential expression of CLCF1 in GBM cancer and non-cancerous samples with the TCGA GBM dataset. As shown in Figure 1A, CLCF1 had significantly higher expression in the cancer group, indicating an oncogenic role of CLCF1 in GBM. Then, we analyzed the prognostic significance of CLCF1 in GBM patients. As shown in Figure 1B,1C, GBM patients with higher CLCF1 expression had a significantly lower overall survival (OS) rate [hazard ratio (HR) =1.5, P value =0.031] and disease-free survival rate (HR =1.8, P value =0.0039), implying that CLCF1 may serve as a risk factor for GBM patients. CGGA datasets were utilized to validate the prognostic significance of CLCF1 in GBM. As shown in Figure 1D, increased expression of CLCF1 was observed in higher WHO grade GBM samples. Additionally, CLCF1 was found to be highly expressed in IDH1 wild-type and non1p/19q co-deletion samples (Figure 1E). Finally, CLCF1 was found to be significantly related to prognosis in primary glioma (P<0.0001), recurrent glioma (P=0.051), grade III (P=0.0022), and IV glioma patients (P=0.011) (Figure 1F).
CLCF1 silencing suppresses cell proliferation, migration, and invasion and enhances apoptosis in GBM in vitro
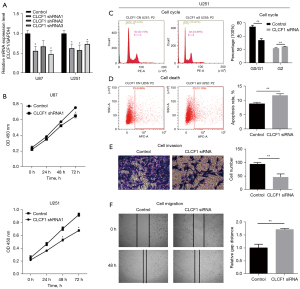
To investigate the biological functions of CLCF1 in GBM U87 and U251 cells, including cell proliferation, migration, invasion, cell cycle, and apoptosis, shRNAs were constructed. Expression of CLCF1 in the CLCF1 knockdown groups was down-regulated compared to that in the control groups (Figure 2A). Cell proliferation in the CLCF1 knockdown groups was found to be significantly lower than that in negative control (NC) in both cell lines (Figure 2B,2C). As CLCF1 silencing exhibited a more pronounced effect in U251 cells, the subsequent assays were conducted in U251 cells. As shown in Figure 2D, CLCF1 knockdown significantly reduced the proportion of cells in G0/G1 phase but slightly induced G2 cell cycle arrest. A higher percentage of apoptotic cells was found in the CLCF1 knockdown groups than in the control groups (Figure 2D). These results suggest that CLCF1 might enhance the cell growth of GBM. The effect of CLCF1 on the migratory and invasive abilities of GBM cells was determined using transwell and wound healing assays, respectively. The transwell invasion assay results indicated that knockdown of CLCF1 reduced cell migration in GBM cells (Figure 2E). Migration in GBM cells was slower in the CLCF1 silencing group than in the control groups (Figure 2F). These results indicate that CLCF1 plays an oncogenic role by promoting cell migration and invasion in vitro.
Related downstream mechanisms of CLCF1 in GBM
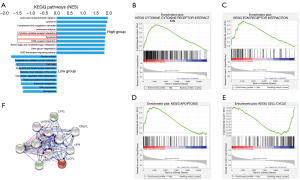
To uncover the molecular mechanism of CLCF1 in GBM, we divided GBM cancer samples into CLCF1 high/low groups and performed GSEA. The top 10 significantly enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in the CLCF1 high and low groups are shown in Figure 3A. For the CLCF1 high group, immune- and cell proliferation-related pathways, such as cytokine-cytokine receptor interaction (Figure 3B), extracellular matrix (ECM) receptor interaction (Figure 3C), and apoptosis (Figure 3D), were significantly enriched. For the CLCF1 low groups, we noticed that the gene set in the cell cycle (Figure 3E) was one of the most significantly enriched terms. To further analyze potential targets of CLCF1, a protein-protein interaction (PPI) network of CLCF1 was constructed using the String database (Figure 3F). According to Figure 3F, CLCF1 was predicted to be related to leukemia inhibitory factor receptor (LIFR), cardiotrophin-1 (CTF1), JAK1, JAK2, signal transducer and activator of transcription 3 (STAT3), CNTF, CNTFR, CRLF1, IL-6 signal transduction (IL6ST), and tyrosine kinase 2 (TYK2). In fact, CLCF1 is a member of the IL-6 cytokine family, and its potential role in inflammatory diseases and cancers has been reported (12). All of these results suggest that CLCF1 may play a complex role in GBM by repressing cell proliferation and regulating the tumor microenvironment through cytokine receptors and ECM receptors.
BRD4 transcriptionally activates CLCF1 in GBM
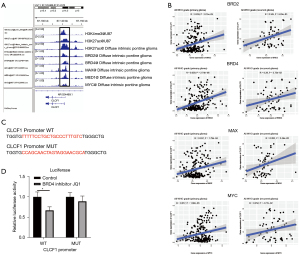
As CLCF1 showed significantly higher expression in GBM, we explored the factors that impact CLCF1 expression. First, we analyzed histone activation marks, such as H3K4me3 and H3K27ac peaks, in glioma cells and samples from the ChIP-Atlas database (24). As shown in Figure 4A, strong binding peak signals were not only found for H3K4me3 and H3K27ac, but also transcription factors such as BRD2, BRD4, MYC, MYC-associated protein X (MAX), and Mediator Subunit 1 (MED1) were found to bind to the CLCF1 promoter. Then, we validated the expression of 5 transcription factors with CLCF1 in the CGGA glioma datasets. As shown in Figure 4B, there was a strong positive correlation coefficient between BRD2, BRD4, MYC, and MAX and CLCF1 in both primary and recurrent glioma samples, suggesting that CLCF1 may be regulated by these transcription factors. As BRD4 showed the highest correlation coefficient with CLCF1 (0.359 for primary glioma and 0.28 for recurrent glioma), we chose BRD4 for subsequent validation. A dual luciferase assay was then conducted to test whether mutation of the BRD4 binding cis-element in the CLCF1 promoter impacts CLCF1 expression. Binding sequencing was mutated, as shown in Figure 4C, and the BRD4 inhibitor JQ1 was used to antagonize BRD4 function. The BRD4 inhibitor JQ1 significantly reduced luciferase activity, while no significant difference was found in the mutant group (Figure 4D), implying that elevated CLCF1 was transcriptionally activated by BRD4 in glioma.
Discussion
Accumulating evidence indicates that cytokines are widely involved in GBM progression and therapy response (6). Previous studies found that the high expression of lncRNA MIR155 host gene (MIR155HG) was a prognostic biomarker and associated with poorer overall survival in glioblastoma multiforme (25). Shugoshin 2 (SGO2) is a biomarker that is predictive of WHO pathological grading and patient survival in patients with gliomas (26). Nicotinamide phosphoribosyltransferase (NAMT), and C1q/TNF-related protein 1 (CTRP1) were reported to be potential prognostic and therapeutic biomarker for glioblastoma (27,28). In this study, we focused on the cytokine CLCF1. We uncovered its clinical significance and function in glioma cells and finally identified potential downstream targets and BRD4 as a transcription activator for the elevated expression of CLCF1 in GBM. By binding to CNTFR, CLCF1 mediates interactions with the co-receptors glycoprotein 130 (gp130) and LIFR. In lung cancer, CLCF1-CNTFR signaling has been observed in cancer-associated fibroblasts (CAFs) and acts to promote cancer cell growth (21). Additionally, a decoy receptor/antagonist (16) engineered to target CLCF1/CNTFR signaling proved effective in inhibiting tumor growth, and the mechanism was related to guanosine triphosphate loading (11). Therefore, this study may provide a new therapeutic target and prognostic marker in GBM.
Given that CLCF1 showed significantly higher expression in GBM cancer samples, we then focused on the CLCF1 downstream pathways and targets to analyze the role of CLCF1 in GBM. As expected, the GSEA pathway analysis identified potential targets in 4 cellular processes: cytokines and receptors, ECM and receptors, apoptosis, and the cell cycle (Figures 3,4). Interestingly, CLCF1 showed positive and negative correlations with pro-apoptosis markers (FAS, BAX, and CASP7) and cell cycle (CCNA2, CCNB1, and CDKN1C) markers, suggesting that CLCF1 may inhibit cancer cell growth in GBM. Furthermore, CLCF1 was positively correlated with cytokines (LIF, CXCL3, and CSF1), cytokine receptors (IL4R, TGFBR2, and CCR1), ECM proteins (COL1A1, COL1A2, and LAMA2), and ECM receptors (ITGA5, ITGA3, and ITGB5), most of which have been reported as oncogenes in GBM (29-31). As cytokines, ECM proteins and their receptors are well-recognized participants in cell-cell communications of the microenvironment (6,32).Their significant correlations with CLCF1 imply that CLCF1 may also play a tumor-promoting role by regulating the GBM microenvironment. In summary, CLCF1 may play a complex role in GBM by repressing cell proliferation and regulating the tumor microenvironment through cytokines and ECM proteins and their respective receptors.
Transcription factors (33) are known regulators of gene expression and are markers in GBM. In this study, we found a strong binding peak and a significant positive correlation between 4 transcription factors and CLCF1, suggesting their transcriptional regulation of CLCF1 expression, and subsequent analysis identified BRD4 as a transcription factor for CLCF1.
BRD4, a member of the bromodomain and extraterminal (BET) protein family, plays an important role in controlling oncogene expression and genome stability (34). In glioma, BRD4 has been reported to promote glioma cell stemness (35-37) and progression (38-41). Recently, studies have indicated that BRD4 is a prospective therapeutic target for the treatment of GBM for its ability to penetrate the blood-brain barrier and target glioma tumor tissues with little side effects (42,43). Therefore, understanding the mechanism of CLCF1 as the target of BRD4 would provide another perspective for the treatment of glioma with BRD4 inhibitors.
Here, this study reveals the expression and prognosis of CLCF1 in glioblastoma based on public information database and bioinformation analysis technology, in order to further clarify the mechanism in the occurrence and development of glioblastoma from a new perspective, seek new effective treatment targets and prolong the survival of patients. Collectively, by integrated bioinformatics analysis, this study revealed CLCF1 as a hazardous prognostic marker in GBM in vitro. CLCF1 might play a complex role in GBM by regulating GBM cancer cells and the immune microenvironment through cytokines, the ECM, and receptors. Finally, BRD4 may be involved in transcriptional activation of CLCF1 in GBM.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1164/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1164/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1164/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Malzkorn B, Reifenberger G. Practical implications of integrated glioma classification according to the World Health Organization classification of tumors of the central nervous system 2016. Curr Opin Oncol 2016;28:494-501. [Crossref] [PubMed]
- Louis DN, Perry A, Wesseling P, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol 2021;23:1231-51. [Crossref] [PubMed]
- Kessler T, Sahm F, Sadik A, et al. Molecular differences in IDH wildtype glioblastoma according to MGMT promoter methylation. Neuro Oncol 2018;20:367-79. [Crossref] [PubMed]
- Jhanwar-Uniyal M, Labagnara M, Friedman M, et al. Glioblastoma: molecular pathways, stem cells and therapeutic targets. Cancers (Basel) 2015;7:538-55. [Crossref] [PubMed]
- Guan R, Zhang X, Guo M. Glioblastoma stem cells and Wnt signaling pathway: molecular mechanisms and therapeutic targets. Chin Neurosurg J 2020;6:25. [Crossref] [PubMed]
- Zhou W, Jiang Z, Li X, et al. Cytokines: shifting the balance between glioma cells and tumor microenvironment after irradiation. J Cancer Res Clin Oncol 2015;141:575-89. [Crossref] [PubMed]
- Manini I, Caponnetto F, Bartolini A, et al. Role of Microenvironment in Glioma Invasion: What We Learned from In Vitro Models. Int J Mol Sci 2018;19:147. [Crossref] [PubMed]
- Lin W, Wu S, Chen X, et al. Characterization of Hypoxia Signature to Evaluate the Tumor Immune Microenvironment and Predict Prognosis in Glioma Groups. Front Oncol 2020;10:796. [Crossref] [PubMed]
- Pitt JM, Marabelle A, Eggermont A, et al. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Ann Oncol 2016;27:1482-92. [Crossref] [PubMed]
- Savin VJ, Sharma M, Zhou J, et al. Renal and Hematological Effects of CLCF-1, a B-Cell-Stimulating Cytokine of the IL-6 Family. J Immunol Res 2015;2015:714964. [Crossref] [PubMed]
- Kim JW, Marquez CP, Kostyrko K, et al. Antitumor activity of an engineered decoy receptor targeting CLCF1-CNTFR signaling in lung adenocarcinoma. Nat Med 2019;25:1783-95. [Crossref] [PubMed]
- Jones SA, Jenkins BJ. Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer. Nat Rev Immunol 2018;18:773-89. [Crossref] [PubMed]
- Nahlé S, Pasquin S, Laplante V, et al. Cardiotrophin-like cytokine (CLCF1) modulates mesenchymal stem cell osteoblastic differentiation. J Biol Chem 2019;294:11952-9. [Crossref] [PubMed]
- Ge JR, Xie LH, Chen J, et al. Liuwei Dihuang Pill () Treats Postmenopausal Osteoporosis with Shen (Kidney) Yin Deficiency via Janus Kinase/Signal Transducer and Activator of Transcription Signal Pathway by Up-regulating Cardiotrophin-Like Cytokine Factor 1 Expression. Chin J Integr Med 2018;24:415-22. [Crossref] [PubMed]
- Pasquin S, Tormo A, Moreau J, et al. Cardiotrophin-Like Cytokine Factor 1 Exhibits a Myeloid-Biased Hematopoietic-Stimulating Function. Front Immunol 2019;10:2133. [Crossref] [PubMed]
- Kim JW, Marquez CP, Sperberg RAP, et al. Engineering a potent receptor superagonist or antagonist from a novel IL-6 family cytokine ligand. Proc Natl Acad Sci U S A 2020;117:14110-8. [Crossref] [PubMed]
- Pasquin S, Laplante V, Kouadri S, et al. Cardiotrophin-like Cytokine Increases Macrophage-Foam Cell Transition. J Immunol 2018;201:2462-71. [Crossref] [PubMed]
- Stefanovic L, Stefanovic B. Role of cytokine receptor-like factor 1 in hepatic stellate cells and fibrosis. World J Hepatol 2012;4:356-64. [Crossref] [PubMed]
- Elsaeidi F, Bemben MA, Zhao XF, et al. Jak/Stat signaling stimulates zebrafish optic nerve regeneration and overcomes the inhibitory actions of Socs3 and Sfpq. J Neurosci 2014;34:2632-44. [Crossref] [PubMed]
- Rousseau F, Gauchat JF, McLeod JG, et al. Inactivation of cardiotrophin-like cytokine, a second ligand for ciliary neurotrophic factor receptor, leads to cold-induced sweating syndrome in a patient. Proc Natl Acad Sci U S A 2006;103:10068-73. [Crossref] [PubMed]
- Vicent S, Sayles LC, Vaka D, et al. Cross-species functional analysis of cancer-associated fibroblasts identifies a critical role for CLCF1 and IL-6 in non-small cell lung cancer in vivo. Cancer Res 2012;72:5744-56. [Crossref] [PubMed]
- Zhang P, Meng X, Liu L, et al. Identification of the Prognostic Signatures of Glioma With Different PTEN Status. Front Oncol 2021;11:633357. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Oki S, Ohta T, Shioi G, et al. ChIP-Atlas: a data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep 2018;19:e46255. [Crossref] [PubMed]
- Peng L, Chen Z, Chen Y, et al. MIR155HG is a prognostic biomarker and associated with immune infiltration and immune checkpoint molecules expression in multiple cancers. Cancer Med 2019;8:7161-73. [Crossref] [PubMed]
- Kao Y, Tsai WC, Chen SH, et al. Shugosin 2 is a biomarker for pathological grading and survival prediction in patients with gliomas. Sci Rep 2021;11:18541. [Crossref] [PubMed]
- Chen L, Su G. Identification of CTRP1 as a Prognostic Biomarker and Oncogene in Human Glioblastoma. Biomed Res Int 2019;2019:2582416. [Crossref] [PubMed]
- Guo Q, Han N, Shi L, et al. NAMPT: A potential prognostic and therapeutic biomarker in patients with glioblastoma. Oncol Rep 2019;42:963-72. [Crossref] [PubMed]
- Chen J, Chen T, Zhu Y, et al. circPTN sponges miR-145-5p/miR-330-5p to promote proliferation and stemness in glioma. J Exp Clin Cancer Res 2019;38:398. [Crossref] [PubMed]
- Chen R, Lee WY, Zhang XH, et al. Epigenetic Modification of the CCL5/CCR1/ERK Axis Enhances Glioma Targeting in Dedifferentiation-Reprogrammed BMSCs. Stem Cell Reports 2017;8:743-57. [Crossref] [PubMed]
- Peñuelas S, Anido J, Prieto-Sánchez RM, et al. TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell 2009;15:315-27. [Crossref] [PubMed]
- Brassart-Pasco S, Brézillon S, Brassart B, et al. Tumor Microenvironment: Extracellular Matrix Alterations Influence Tumor Progression. Front Oncol 2020;10:397. [Crossref] [PubMed]
- Sen E. Targeting inflammation-induced transcription factor activation: an open frontier for glioma therapy. Drug Discov Today 2011;16:1044-51. [Crossref] [PubMed]
- Du Z, Song X, Yan F, et al. Genome-wide transcriptional analysis of BRD4-regulated genes and pathways in human glioma U251 cells. Int J Oncol 2018;52:1415-26. [Crossref] [PubMed]
- Wang J, Quan Y, Lv J, et al. BRD4 promotes glioma cell stemness via enhancing miR-142-5p-mediated activation of Wnt/beta-catenin signaling. Environ Toxicol 2020;35:368-76. [Crossref] [PubMed]
- Tao Z, Li X, Wang H, et al. BRD4 regulates self-renewal ability and tumorigenicity of glioma-initiating cells by enrichment in the Notch1 promoter region. Clin Transl Med 2020;10:e181. [Crossref] [PubMed]
- Wang Q, Jia S, Wang D, et al. A Combination of BRD4 and HDAC3 Inhibitors Synergistically Suppresses Glioma Stem Cell Growth by Blocking GLI1/IL6/STAT3 Signaling Axis. Mol Cancer Ther 2020;19:2542-53. [Crossref] [PubMed]
- Wen N, Guo B, Zheng H, et al. Bromodomain inhibitor jq1 induces cell cycle arrest and apoptosis of glioma stem cells through the VEGF/PI3K/AKT signaling pathway. Int J Oncol 2019;55:879-95. [Crossref] [PubMed]
- Wang Q, Kumar V, Lin F, et al. ApoE mimetic peptide targeted nanoparticles carrying a BRD4 inhibitor for treating Medulloblastoma in mice. J Control Release 2020;323:463-74. [Crossref] [PubMed]
- Pastori C, Kapranov P, Penas C, et al. The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc Natl Acad Sci U S A 2015;112:8326-31. [Crossref] [PubMed]
- Xu L, Chen Y, Mayakonda A, et al. Targetable BET proteins- and E2F1-dependent transcriptional program maintains the malignancy of glioblastoma. Proc Natl Acad Sci U S A 2018;115:E5086-95. [Crossref] [PubMed]
- Yang H, Wei L, Xun Y, et al. BRD4: An emerging prospective therapeutic target in glioma. Mol Ther Oncolytics 2021;21:1-14. [Crossref] [PubMed]
- Kfoury N, Qi Z, Prager BC, et al. Brd4-bound enhancers drive cell-intrinsic sex differences in glioblastoma. Proc Natl Acad Sci U S A 2021;118:e2017148118. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


