
This article has an erratum available at: http://dx.doi.org/10.21037/atm-2024-13 the article has been update on 2024-08-07 at here.
Transthyretin-induced increase in circ_0007411 represses neovascularization of human retinal microvascular endothelial cells in hyperglycemia via the miR-548m/PTPN12/SKP1/EGFR pathway
Introduction
In response to the dramatically increasing prevalence of diabetes (1), diabetic retinopathy (DR) has been identified as a major cause of blindness and visual impairment (2,3). During diabetes, the sustained hyperglycemia and the subsequent ischemic retinal hypoxia have been considered as the important triggers for vascular dysfunction; and the downstream switches, such as the hypoxia-inducible factor-1α (HIF-1α) or vascular endothelial growth factor (VEGF) associated pathways, would be stimulated to promote the progression of DR, bringing with the clinical properties of DR, including microaneurysms, hemorrhages, lipid exudates, diabetic macular edema (DME), retinal capillary occlusion, cotton-wool spots, and neovascularization (NV) (3,4). In addition, as recently reported, inflammation associated pathways have also been considered as the key factors in the progression of DR, including nuclear factor-κB (NF-κB), Toll-like receptor (TLR) and Jak/Stat pathways; and the chronic low-grade inflammation in retina could promote the edema and NV in DR (5).
The prevention of retinal NV has recently been considered as the basic principle for the clinical therapy of DR, and to rescue retinal vascular leakage and angiogenesis in DR, anti-VEGF antibodies, including pegaptanib, ranibizumab, and bevacizumab, have been applied through intraocular injection (6). However, the development of more efficient therapy protocols requires systematic investigation into the mechanism of retinal microvascularization.
Circular RNAs (circRNAs) are the back-splicing products after transcription, without 5'–3' polarity and poly A tails, circRNAs are more stable than mRNAs, and they have been proved as sponges of micro-RNAs, or to interact with RNA binding proteins, regulate mRNAs stability, modulate gene transcription and translate proteins (7). And recently, circRNAs have emerged as a hot topic in the study of various diseases, including diabetes and eye disease (8,9), some of them have been demonstrated to play vital roles in the procession of DR by regulating the angiogenesis, proliferation, apoptosis, and inflammatory response in the retina of DR patients and animal models (10). In addition, some circRNAs have been reported to affect the physiological properties of retinal microvascular endothelial cells, including circRNA-FoxO1 (11), circHIPK3 (12), and circCOL1A2 (13). These studies suggest circRNAs may be potential biomarkers for the diagnosis of DR or molecular targets clinical therapy.
Protein tyrosine phosphatase nonreceptor type 12 (PTPN12) is located at chromosome 7q11.23 (14) and belongs to the protein tyrosine phosphatase family. PTPN12 has been reported to participate in tumor occurrence and development (15), embryonic development, cell apoptosis, cell cycle, and metabolism (16), and has also been reported to exert antitumor function by regulating epidermal growth factor receptor (EGFR) (17). Circ_0007411 is one circRNA form of PTPN12, but its role in the mechanism of NV has rarely been investigated.
In our previous work, transthyretin (TTR) was shown to have an anti-angiogenic function in DR, mediated through the PABPC1/lncRNA MEG3/miR-223-3p/FBXW7 and hnRNPA2B1/STAT4/miR-223-3p/FBXW7 signaling axes (18-20). However, it is not known if TTR can affect or regulate circRNAs. In this study, after human retinal endothelial cells (hRECs) was treated with TTR, abnormally expressed circRNAs were screened via RNA sequencing (RNA-Seq), and circ_0007411 was significantly promoted in hyperglycemia with exogenous TTR. Circ_0007411 has not been reported as associated with the progression of DR; in addition, bioinformatics assay indicated that circ_0007411 contained the same target sites of miR-548m in the 3'-untranslated region (UTR) of PTPN12 mRNA. The relationship between circ_0007411, PTPN12, and miR-548m was investigated in hRECs, and the anti-NV function of PTPN12 was investigated in a STZ-induced model of DR in C57BL/6 mice. The mechanisms were also studied. We present the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1276/rc).
Methods
Cell culture and reagents
hRECs were obtained from the Cell Bank of the Chinese Academy of Science (Shanghai, China), using high-glucose DMEM (Gibco, USA) containing 10% FBS for cultivation, as described in our previous work (15-17). Anti-PTPN12, -SKP1, -EGFR, and -glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA), while the quantitative real-time polymerase chain reaction (qRT-PCR) primers for circ_0007411, PTPN12, miR-548m, cel-miR-39-3p, and GAPDH were synthesized by GenePharma (Suzhou, China). The mouse PTPN12 overexpression adeno-associated virus (AAV), shPTPN12 (5'-GGACATACTGCCATTTGATCACGAATGATCAAATGGCAGTATGTCC-3'), circ_0007411 overexpression plasmid (OECIRC), si-circ_0007411-1 (5'-GUAUUCAUUGCAGUUGAUCTT-3'), si-circ_0007411-2 (5'-CAUUGCAGUUGAUCACAGCTT-3'), and the miR-548m mimic and inhibitor were synthesized by GenePharma (Suzhou, China). The cells were grouped as high glucose (HG), overexpression-negative control (OE-NC), OE-7411, si-NC, si-7411-1, si-7411-2, mimic-NC, miR-548m mimic, mimic + 7411, inhibitor NC, and miR-548m inhibitor; and each experiment was repeated for at least three times.
RNA-Seq
Trizol was used to extract total RNA from hRECs cultivated in a high-glucose environment with or without 4 μmol/L TTR in accordance with the manufacturer’s handbook (Takara, Dalian, China). After the RNA concentration was detected with a Qubit RNA detection kit (Invitrogen, Carlsbad, CA, USA), the extracted RNA was subjected to sequencing, and the raw data were deposited in the Gene Expression Omnibus-National Center for Biological Information (GEO-NCBI; GSE117238) (21). After screening using FastQC (V0.11.9) and NGSQC software (V2.3.3) (22), circRNAs were further identified by Tophat2 (v2.1.1), Find_circ (v1.2), and CIRCexplorer2 software (23), and differentially expressed circRNAs were identified using the Limma (v3.44.3) software package (24).
Cell migration and healing
As described in our previous work (18-20), hRECs were inoculated in the Transwell insert, incubated for 30 h, fixed with 4% paraformaldehyde, stained with crystal violet, and counted under light microscope. Using a wound healing assay kit (ab242285), the hRECs were plated in six-well culture dishes and grown to confluence. A defined gap was then created by the insert of the kit, and wound healing was monitored over 24 h. The relative wound density (RWD) was detected and calculated using an Olympus IX-73 microscope.
Tube formation
After a 0.5 h incubation at 37 ℃ in in a 48-well plate, the hRECs were inoculated into the basement membrane matrix (BD Biosciences) for a further 3 h incubation at 37 ℃, and the tube formation process was observed and monitored using an Olympus IX-73 microscope (18-20).
qRT-PCR
As described in our previous work (18-20), after extraction of the total RNAs and total miRNAs, the expression of PTPN12 mRNA and circ_0007411 in hRECs was detected using Moloney murine leukemia virus (M-MLV) reverse transcriptase and a SYBR Green Real-Time PCR Master Mix (Invitrogen, Carlsbad, CA, USA), with GAPDH was as the internal control. The level of miR-548M was detected using the mirVanaTM qRT-PCR miRNA detection kit (Invitrogen, Carlsbad, CA, USA), with cel-miR-39-3p spike-in as the internal control. The primers are listed in Table 1.
Table 1
| Target gene | Primers |
|---|---|
| circ_0007411 | F: 5'-GCCAGACCATGATGTTCCTTC-3' |
| R: 5'-TGCTAAAGGTCCTTGAGTTGC-3' | |
| PTPN12 | F: 5'-CTCCTCCCCTACCTGAAAG-3' |
| R: 5’-TTCACTTGCTAACACAAACGA-3' | |
| SKP1 | F: 5'-GACCATGTTGGAAGATTTGGGA-3' |
| R: 5'-TGCACCACTGAATGACCTTTT-3' | |
| EGFR | F: 5'-CTACAACCCCACCACGTACC-3' |
| R: 5'-CGCACTTCTTACACTTGCGG-3' | |
| miR-548m | F: 5'-AAGCGACCCAAAGGTATTTGT-3' |
| R: 5'-GTCGTATCCAGTGCAGGGT-3' | |
| cel-miR-39-3p | F: 5'-GTCACCGGGTGTAAATCAG-3' |
| R: 5'-GGTCCAGTTTTTTTTTTTTTTTCAAG-3' | |
| GAPDH | F: 5'-GCACCGTCAAGGCTGAGAAC-3' |
| R: 5'-TGGTGAAGACGCCAGTGGA-3' |
qRT-PCR, quantitative real-time polymerase chain reaction; PTPN12, protein tyrosine phosphatase nonreceptor type 12; SKP1, S-phase kinase associated protein 1; EGFR, epidermal growth factor receptor; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Flow cytometry (FCM)
Apoptotic cells were labelled using the Annexin V/FITC kit (BD Biosciences, San Jose, CA, USA) in accordance with the manufacturer’s instructions, and monitored by FCM.
Cell Counting Kit-8 (CCK-8) assay
The proliferation of hRECs was investigated using a CCK-8 kit (Dojindo Laboratories, Kumamoto, Japan) as described in our previous work (18,21).
5-Ethynyl-2′-deoxyuridine (EdU)
The EdU assay was conducted in accordance with the manufacturer’s handbook (RiboBio, Guangzhou, China). After hRECs were seeded in a 48-well plate, 50 μM EdU was added to each well, and the plate was incubated at 37 ℃ for 4 h. The cells were fixed in 4% paraformaldehyde and permeabilized by 0.5% Triton X-100 then sequentially treated with Apollo 488 fluorescent dye solution and 4',6-diamidino-2-phenylindole (DAPI) solution. The image was captured using a fluorescence confocal microscope (Zeiss, Oberkochen, Germany).
Dual-luciferase reporter
Wild or mutated circ_0007411 or PTPN12 3'-UTR were synthesized into pGL3-luc plasmids containing luciferase reporter gene together with miR-548m (GenePharma, Shanghai, China). The plasmids were transiently transfected into hRECs using the Lipofectamine 3000 transfection system, and then the Dual Luciferase Assay System (Promega, Madison, WI, USA) was employed to test the firefly luciferase activity and Renilla activity.
Co-immunoprecipitation (Co-IP) and Western blotting
PTPN12 and SKP1 antibodies were added to extracts of lysed hRECs, respectively, then incubated for 12 h at 4 ℃. Sepharose beads coupled with Protein A were then added to the mixture and incubated for a further 4 h. The beads were washed twice with PBS and three times by washing buffer. After boiling with 50 μL of 1× sodium dodecyl sulfate (SDS) loading buffer for 10 min, the proteins were eluted then subjected to Western blotting assay as described in our previous work (18-20).
STZ-induced DR mouse model
Animal experiments were performed under a project license (No. 2019-398) granted by the Ethics Committee of Nanjing Medical University, in compliance with institutional guidelines for the humane treatment of animals, the Principles of Laboratory Animal Care (National Institutes of Health, Bethesda, MD, USA) (https://www.ncbi.nlm.nih.gov/books/NBK54050), and the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research (https://www.arvo.org/About/policies/statement-for-the-use-of-animals-in-ophthalmic-and-vision-research/).
A total of 50 μg/g STZ was injected intraperitoneally into 8-week-old male C57BL/6 mice (purchased from Changzhou Cavens Laboratory Animals Co., Ltd.) for 5 days continuously. After an over 16.7 mmol/L blood glucose level (19), 80 μg/g ketamine was intraperitoneally injected together with 4 μg/g xylazine, and then the mice received an intravitreal injection of ~1.5 μL (1×1012 TU/mL) of AAV carrying mouse PTPN12 complementary DNA (cDNA) every 4 weeks. For shPTPN12 (5'-CACCGGACATACTGCCATTTGATCACGAATGATCAAATGGCAGTATGTCC-3'), the mice were administered an intravitreal injection (2 nmoL) every 2 weeks. The animals were grouped as: normal, STZ, STZ NC, STZ + OE-PTPN12, STZ + shPTPN12, with 5 mice in each group. After approximately 16 weeks, the retinal was recovered and investigated using Evans blue leakage and retinal trypsin digestion assays in our previous work (19). The eyeballs were enucleated and fixed with 4% paraformaldehyde for 24 h, and the retina and sclera were embedded in paraffin after dehydration. In hematoxylin and eosin (H&E) staining, 5-μm-thick sections were taken along the vertical meridian and were visualized using an Olympus BX-51 light microscope.
Statistical analysis
All in vitro experiments were repeated at least three times and all data were expressed as the average ± standard deviation (SD). T-test and one-way ANOVA were employed for comparisons between two groups or multiple groups in SPSS software (v 13.0). A P value of <0.05 was considered statistically significant.
Results
TTR-induced high level of circ_0007411 prevented the proliferation, migration, wound healing, and tube formation of hRECs in a high-glucose environment
In RNA-Seq and after TTR treatment, 78 differentially expressed circRNAs were identified, and circ_0007411 derived from PTPN12 was significantly upregulated (Figure 1A). qRT-PCR analysis indicated that in a high-glucose environment, the level of circ_0007411 was increased by the addition of TTR (Figure 1B). CCK-8 assay (Table 2) and EdU labelling (Figure 1C,1D) suggested the overexpression of circ_0007411 inhibited the proliferation of hRECs, whereas treatment with the two siRNAs promoted the proliferation. In FCM assay, the overexpression of circ_0007411 promoted the apoptosis ratio, whereas the siRNAs resulted in no significant differences (Figure 1E,1F). In Transwell assay, the migration of hRECs was repressed by overexpression of circ_0007411, but the siRNAs caused an increase (Figure 1G). Additionally, wound healing (Figure 1H,1I) and tube formation (Figure 1J,1K) were also suppressed by a high level of circ_0007411.
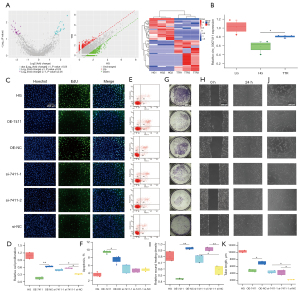
Table 2
| Groups | 24 h | 48 h | 72 h | |||||
|---|---|---|---|---|---|---|---|---|
| OD450nm, mean ± SD | P value | OD450nm, mean ± SD | P value | OD450nm, mean ± SD | P value | |||
| HG | 1.947±0.223 | 2.985±0.302 | 3.655±0.344 | |||||
| OE-7411 | 1.235±0.083 | OE-7411 vs. OE-NC, 0.086 | 2.107±0.124 | OE-7411 vs. OE-NC, 0.030 | 2.860±0.130 | OE-7411 vs. OE-NC, 0.042 | ||
| OE-NC | 1.458±0.088 | 2.536±0.188 | 3.295±0.121 | |||||
| si-7411-1 | 1.424±0.074 | si-7411-1 vs. si-NC, 0.715; si-7411-2 vs. si-NC, 0.278 | 2.617±0.156 | si-7411-1 vs. si-NC, 0.042; si-7411-2 vs. si-NC, 0.028 | 3.331±0.160 | si-7411-1 vs. si-NC, 0.033; si-7411-2 vs. si-NC, 0.040 | ||
| si-7411-2 | 1.502±0.093 | 2.589±0.123 | 3.264±0.122 | |||||
| si-NC | 1.385±0.088 | 2.190±0.070 | 2.793±0.147 | |||||
HG, high glucose; OE, overexpression; NC, negative control; hRECs, human retinal microvascular endothelial cells; OD, optical density; SD, standard deviation.
Circ_0007411 regulated the level of PTPN12 by targeting miR-548m
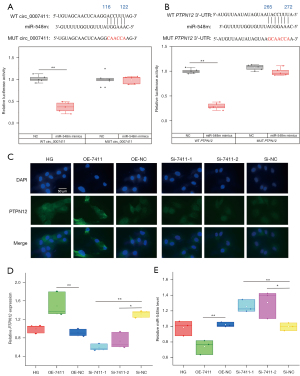
The NIH Circular RNA Interactome online service (https://circinteractome.nia.nih.gov/mirna_target_sites.html) and the TargetScanHuman online service (http://www.targetscan.org/vert_70/) were employed to screen the possible targets of circ_0007411 and the 3'-untranslation region (UTR) of PTPN12. The results suggested circ_0007411 [116–122] (Figure 2A) and the 3'-UTR of PTPN12 mRNA [265–272] (Figure 2B) shared similar binding positions of miR-548m. To investigate whether miR-548m could bind with circ_0007411/PTPN12 mRNA, the wild-type (WT) and mutant (MUT) sequences containing the candidate sites were transfected into hRECs, respectively, and the transient co-transfection with plasmids containing WT sites and miR-548m mimics showed decreased luciferase activities (Figure 2A,2B). In addition, the protein (Figure 2C) and mRNA (Figure 2D) levels of PTPN12 were increased in hRECs after the overexpression of circ_0007411 but were suppressed by the knockdown of circ_0007411. In contrast, the level of miR-548m was repressed after the overexpression of circ_0007411 but was enhanced by the two siRNAs (Figure 2E).
Circ_0007411 might affect hRECs via the miR-548m/PTPN12/SKP1/EGFR pathway
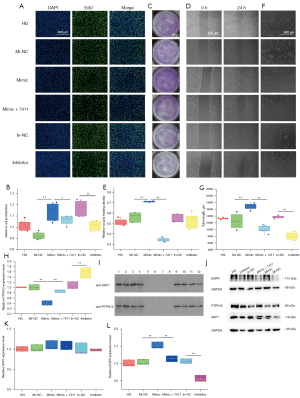
To evaluate whether miR-548m could affect the hRECs in a high-glucose environment and to determine whether circ_0007411 could reverse these phenomena, miR-548m mimic, miR-548m inhibitor, and circ_0007411 overexpression plasmid (OECIRC) were co-transfected into hRECs. CCK-8 assay (Table 3), EdU labelling (Figure 3A,3B), Transwell (Figure 3C), wound healing (Figure 3D,3E), and tube formation (Figure 3F,3G) assays indicated miR-548m mimics could significantly promote the proliferation, migration, wound healing, and tube formation of hRECs, whereas the overexpression of circ_0007411 caused similar effects to the inhibitor of miR-548m and could partially rescue these processes. In addition, miR-548m mimics could significantly reduce the expression of PTPN12, although this phenomenon was rescued by the overexpression of circ_0007411 (Figure 3H). It has been previously reported that PTPN12 plays vital roles in tumor occurrence and development, cell apoptosis, cell cycle, and metabolism and may regulate the content of SKP1 and EGFR (12-14). In this study, the direct binding between PTPN12 and SKP1 was confirmed by co-IP assay (Figure 3I), and Western blotting analysis suggested the protein content of PTPN12, SKP1, and EGFR could be regulated by miR-548m mimic and rescued by circ_0007411 overexpression (Figure 3J). Interestingly, miR-548m mimic could not affect the expression of SKP1 (Figure 3K), although it enhanced the expression of EGFR and was partially rescued by overexpression of circ_0007411 (Figure 3L).
Table 3
| Groups | 24 h | 48 h | 72 h | |||||
|---|---|---|---|---|---|---|---|---|
| OD450nm, mean ± SD | P value | OD450nm, mean ± SD | P value | OD450nm, mean ± SD | P value | |||
| HG | 2.021±0.176 | 3.028±0.272 | 3.812±0.197 | |||||
| Mi-NC | 1.833±0.140 | Mimic vs. mi-NC, 0.014; mimic vs. mimic + 7411, 0.039 | 2.122±0.083 | Mimic vs. mi-NC, 0.004; mimic vs. mimic + 7411, 0.009 | 2.993±0.103 | Mimic vs. mi-NC, 0.008; mimic vs. mimic + 7411, 0.005 | ||
| Mimic | 2.437±0.109 | 2.730±0.101 | 3.455±0.107 | |||||
| Mimic + 7411 | 2.107±0.074 | 2.187±0.112 | 2.915±0.111 | |||||
| In-NC | 1.554±0.082 | In-NC vs. inhibitor, 0.108 | 2.776±0.084 | In-NC vs. inhibitor, 0.031 | 3.032±0.121 | In-NC vs. inhibitor, 0.025 | ||
| Inhibitor | 1.389±0.060 | 2.333±0.150 | 2.560±0.110 | |||||
HG, high glucose; hRECs, human retinal microvascular endothelial cells; OD, optical density; mi-NC, mimic-negative control; in-NC, inhibitor-negative control; SD, standard deviation.
The anti-retinal NV function of PTPN12 in DR mice
As circ_0007411 and miR-548m are not conserved in the mouse, in this work, only the overexpression and knockdown of PTPN12 were performed in the DR mouse model to investigate the effects in vivo. Regarding Figure 4A,4B, the vascular leakage of retina in normal mice (10.1%±3.4%, 4/10 eyes) was not as severe as that in the STZ-treated DR mice (32.5%±4.7%, 4/10 eyes) and the STZ-treated NC mice (28.6%±1.9%, 4/10 eyes). The overexpression of PTPN12 (12.2%±1.3%, 4/10 eyes) could partially rescue the leakage ratio, whereas the knockdown of PTPN12 (44.6%±5.7%, 4/10 eyes) resulted in continuing deterioration.
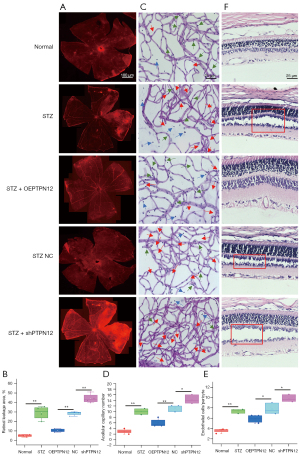
Digested using trypsin, the average number of retinal acellular capillaries (10 fields) was significantly increased from 3±1 of normal mice (4/10 eyes) to 10±2 of STZ-treated DR mice (4/10 eyes) and 11±2 of STZ-treated NC mice (4/10 eyes) but was partially rescued together with the overexpression of PTPN12 (11±2, 4/10 eyes), whereas the knockdown of PTPN12 resulted in continuing deterioration (15±2, 4/10 eyes) (Figure 4C,4D). In addition, as shown in Figure 4C,4E, the hRECs:pericytes ratio (3.2±0.3, 4/10 eyes) was significantly increased in STZ-treated DR mice (7.1±0.8, 4/10 eyes) and STZ-treated NC mice (7.9±1.0, 4/10 eyes), which was associated with the loss of pericytes. The overexpression of PTPN12 partially reversed the loss of pericytes (5.7±0.7, 4/10 eyes), whereas the knockdown of PTPN12 resulted in continuing deterioration (10.2±1.1, 4/10 eyes). Furthermore, the detachment of the retina and choroid in STZ-treated DR mice was partially rescued by overexpression of PTPN12 but was much more severe after the knockdown of PTPN12 (Figure 4F).
Discussion
Diabetes has recently emerged as a global health risk, and DR is one of the most severe complications (2,3). TTR is a 55-kDa homo-tetramer protein which is normally known as the carrier protein of thyroxine (T4) and retinol in plasma and cerebrospinal fluid. TTR has been proven to have anti-retinal NV function in DR in vitro and in vivo through the TTR/PABPC1/lncRNA MEG3/miR-223-3p/FBXW7 and TTR/hnRNPA2B1/STAT4/miR-223-3p/FBXW7 signaling axis (18-20). CircRNAs have been found to play important roles in the mechanism of different diseases, including DR (7-10).
In this work, to screen potential circRNAs that may have been regulated by TTR in hypoglycemia, RNA-Seq was employed. Circ_0007411 from PTPN12 was seen to be promoted by TTR in a high-glucose environment and the mechanism is thought to be associated with the formation of the TTR-hnRNPA2B1 complex, because hnRNPA2B1 has been shown to have mRNA editing activity (20). The overexpression of circ_0007411 could significantly repress the proliferation, migration, wound healing, and tube formation of hRECs in a high-glucose environment, and could promote the proportion of cells in undergoing apoptosis. Another interesting result was that the expression of PTPN12, the parent gene of circ_0007411, was enhanced at both the mRNA and protein levels. PTPN12 has been reported to exert antitumor effects by regulating EGFR (15-17), and SKP1 has been reported to participate in cancer development, and is predicted to regulate EGFR (25,26). However, the details of the mechanism are still unclear.
Predicted using NIH Circular RNA Interactome (https://circinteractome.nia.nih.gov/mirna_target_sites.html) and TargetScanHuman (http://www.targetscan.org/vert_70/), miR-548m was the binding target of circ_0007411 and the 3'-UTR of PTPN12 mRNA, and the direct interaction was confirmed by dual-luciferase reporter assay. In the qRT-PCR assay, the overexpression of circ_0007411 significantly promoted the expression of PTPN12, whereas miR-548m mimics repressed it, and overexpression of circ_0007411 rescued the effects of miR-548m. In addition, in the physiological assays, the proliferation, migration, wound healing, and tube formation progressions were significantly promoted with miR-548m mimics, while these phenomena could be partially rescued by overexpressed circ_0007411. These results suggested that a high level of circ_0007411 could promote the expression of PTPN12 via repression of the miR-548m. Although miR-538m has been reported to suppress the migration and invasion of breast cancer cells (27), it may perform the opposite function in hRECs. We speculated there was a potential interaction between PTPN12 and SKP1 and confirmed the direct binding of PTPN12 and SKP1 in a co-IP assay. Moreover, in a Western blotting assay, miR-548m mimics could regulate the protein levels of PTPN12, SKP1 and EGFR, while the overexpression of circ_0007411 could rescue it. Further, in qRT-PCR assay, miR-548m could only affect the expression of EGFR but not SKP1. These results suggested SKP1 is a direct downstream molecule of PTPN12, and that EGFR is a potential downstream factor of the PTPN12-SKP1 complex, although further investigation is required to determine the details. Additionally, as inflammation has been considered as a central role player in DR, and in our previous work, the antagonistic mechanism of miR-1243 against RELB/circ_0008590 in NF-κB non-classic pathway was proved to repress the NV progression of DR (28); in this work, we tried to investigate the relationship between circ_0007411 and classic inflammation signaling pathways, however, the overexpression and knockdown of circ_0007411 could not affect the factors in NF-κB, TLR and Jak/Stat pathways.
As circ_0007411 and miR-548m are not conserved in mice, in this work, the function of PTPN12 was investigated by establishing an STZ-induced model of DR in C57BL/6 mice. The in vivo results were in good accordance with the in vitro results in hRECs, and the high expression of PTPN12 significantly reduced the procession of DR (29), while the retinal vascular leakage, acellular capillary, loss of pericyte, and the detachment of retina and choroid were all significantly rescued.
Recently, intraocular injection of anti-VEGF antibodies has been considered as the most frequently used clinical therapy for DR, and circ_0007411 should be of great potential as a concomitant candidate. The results of this work suggested that circ_0007411 could relieve the clinical progression of DR through a different pathway together with anti-VEGF therapy. However, owing to the stable and tight ocular barriers (30), how to deliver circ_0007411 into the eye and to keep a stable level in retinal are still critical challenges; more delivering and sustained releasing protocols should be investigated.
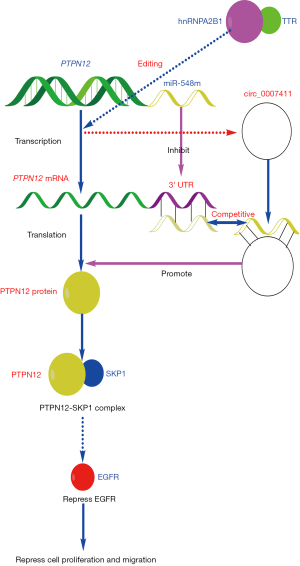
In summary, this was the first study to investigate the TTR-induced high level of circ_0007411 and its downstream mechanism. The identified interaction between circ_0007411, PTPN12 mRNA, and miR-548m, and the direct binding of PTPN12 and SKP1 suggested that circ_0007411 plays a significant role in the anti-angiogenesis process via the miR-548m/PTPN12/SKP1/EGFR signaling pathway (Figure 5).
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 81970819), China Postdoctoral Science Foundation (No. 2020M671541), and Youth Medical Talent Project of Jiangsu Province (No. QNRC2016182).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1276/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-1276/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-1276/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Animal experiments were performed under a project license (No. 2019-398) granted by the Ethics Committee of Nanjing Medical University, in compliance with institutional guidelines for the humane treatment of animals, the Principles of Laboratory Animal Care (National Institutes of Health, Bethesda, MD, USA), and the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wong TY, Sabanayagam C. Strategies to Tackle the Global Burden of Diabetic Retinopathy: From Epidemiology to Artificial Intelligence. Ophthalmologica 2020;243:9-20. [Crossref] [PubMed]
- Amoaku WM, Ghanchi F, Bailey C, et al. Diabetic retinopathy and diabetic macular oedema pathways and management: UK Consensus Working Group. Eye (Lond) 2020;34:1-51. [Crossref] [PubMed]
- Ting DS, Cheung GC, Wong TY. Diabetic retinopathy: global prevalence, major risk factors, screening practices and public health challenges: a review. Clin Exp Ophthalmol 2016;44:260-77. [Crossref] [PubMed]
- Klein R, Lee KE, Gangnon RE, et al. The 25-year incidence of visual impairment in type 1 diabetes mellitus the wisconsin epidemiologic study of diabetic retinopathy. Ophthalmology 2010;117:63-70. [Crossref] [PubMed]
- Capitão M, Soares R. Angiogenesis and Inflammation Crosstalk in Diabetic Retinopathy. J Cell Biochem 2016;117:2443-53. [Crossref] [PubMed]
- Lu L, Jiang Y, Jaganathan R, et al. Current Advances in Pharmacotherapy and Technology for Diabetic Retinopathy: A Systematic Review. J Ophthalmol 2018;2018:1694187. [PubMed]
- Zhou HR, Kuang HY. Circular RNAs: Novel target of diabetic retinopathy. Rev Endocr Metab Disord 2021;22:205-16. [Crossref] [PubMed]
- Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol 2020;21:475-90. [Crossref] [PubMed]
- Guo N, Liu XF, Pant OP, et al. Circular RNAs: Novel Promising Biomarkers in Ocular Diseases. Int J Med Sci 2019;16:513-8. [Crossref] [PubMed]
- He M, Zhou R, Liu S, et al. Circular RNAs: Potential Star Molecules Involved in Diabetic Retinopathy. Curr Eye Res 2021;46:277-83. [Crossref] [PubMed]
- Li D, Liu C, Sun YN, et al. Targeting choroidal vascular dysfunction via inhibition of circRNA-FoxO1 for prevention and management of myopic pathology. Mol Ther 2021;29:2268-80. [Crossref] [PubMed]
- Shan K, Liu C, Liu BH, et al. Circular Noncoding RNA HIPK3 Mediates Retinal Vascular Dysfunction in Diabetes Mellitus. Circulation 2017;136:1629-42. [Crossref] [PubMed]
- Zou J, Liu KC, Wang WP, et al. Circular RNA COL1A2 promotes angiogenesis via regulating miR-29b/VEGF axis in diabetic retinopathy. Life Sci 2020;256:117888. [Crossref] [PubMed]
- Lee C, Rhee I. Important roles of protein tyrosine phosphatase PTPN12 in tumor progression. Pharmacol Res 2019;144:73-8. [Crossref] [PubMed]
- Li H, Yang F, Liu C, et al. Crystal Structure and Substrate Specificity of PTPN12. Cell Rep 2016;15:1345-58. [Crossref] [PubMed]
- de Voer RM, Hahn MM, Weren RD, et al. Identification of Novel Candidate Genes for Early-Onset Colorectal Cancer Susceptibility. PLoS Genet 2016;12:e1005880. [Crossref] [PubMed]
- Lin Q, Wang H, Lin X, et al. PTPN12 Affects Nasopharyngeal Carcinoma Cell Proliferation and Migration Through Regulating EGFR. Cancer Biother Radiopharm 2018;33:60-4. [Crossref] [PubMed]
- Shao J, Fan G, Yin X, et al. A novel transthyretin/STAT4/miR-223-3p/FBXW7 signaling pathway affects neovascularization in diabetic retinopathy. Mol Cell Endocrinol 2019;498:110541. [Crossref] [PubMed]
- Fan G, Gu Y, Zhang J, et al. Transthyretin Upregulates Long Non-Coding RNA MEG3 by Affecting PABPC1 in Diabetic Retinopathy. Int J Mol Sci 2019;20:6313. [Crossref] [PubMed]
- Gu Y, Hu D, Xin Y, et al. Transthyretin affects the proliferation and migration of human retinal microvascular endothelial cells in hyperglycemia via hnRNPA2B1. Biochem Biophys Res Commun 2021;557:280-7. [Crossref] [PubMed]
- Shao J, Zhang Y, Fan G, et al. Transcriptome analysis identified a novel 3-LncRNA regulatory network of transthyretin attenuating glucose induced hRECs dysfunction in diabetic retinopathy. BMC Med Genomics 2019;12:134. [Crossref] [PubMed]
- Dai M, Thompson RC, Maher C, et al. NGSQC: cross-platform quality analysis pipeline for deep sequencing data. BMC Genomics 2010;11:S7. [Crossref] [PubMed]
- Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013;14:R36. [Crossref] [PubMed]
- Zhang XO, Wang HB, Zhang Y, et al. Complementary sequence-mediated exon circularization. Cell 2014;159:134-47. [Crossref] [PubMed]
- Mertins P, Mani DR, Ruggles KV, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 2016;534:55-62. [Crossref] [PubMed]
- Tian C, Lang T, Qiu J, et al. SKP1 promotes YAP-mediated colorectal cancer stemness via suppressing RASSF1. Cancer Cell Int 2020;20:579. [Crossref] [PubMed]
- Wm Nor WMFSB, Chung I, Said NABM. MicroRNA-548m Suppresses Cell Migration and Invasion by Targeting Aryl Hydrocarbon Receptor in Breast Cancer Cells. Oncol Res 2021;28:615-29. [Crossref] [PubMed]
- Shao J, Cai J, Yao Y, et al. Hyperglycemia-induced increasing of RELB/circ_0008590 in NF-κB pathway is repressed by miR-1243 in human retinal microvascular endothelial cells. Ann Transl Med 2021;9:1624. [Crossref] [PubMed]
- Kur J, Newman EA, Chan-Ling T. Cellular and physiological mechanisms underlying blood flow regulation in the retina and choroid in health and disease. Prog Retin Eye Res 2012;31:377-406. [Crossref] [PubMed]
- Lee J, Pelis RM. Drug Transport by the Blood-Aqueous Humor Barrier of the Eye. Drug Metab Dispos 2016;44:1675-81. [Crossref] [PubMed]
(English Language Editor: B. Draper)


